
Lake Sarah-associated circular virus-51
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
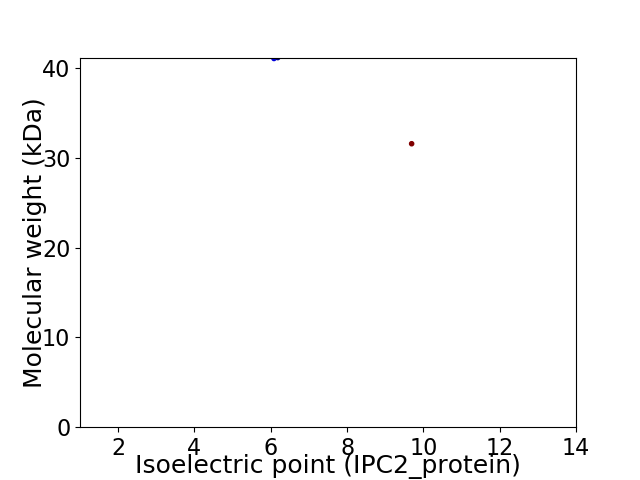
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126GA35|A0A126GA35_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-51 OX=1685781 PE=3 SV=1
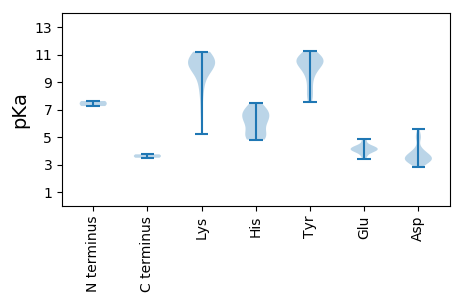
MM1 pKa = 7.25QNTHH5 pKa = 6.46ASSDD9 pKa = 3.91EE10 pKa = 4.05EE11 pKa = 4.63VEE13 pKa = 4.3AGPSRR18 pKa = 11.84TFRR21 pKa = 11.84IDD23 pKa = 3.11ARR25 pKa = 11.84HH26 pKa = 6.5LEE28 pKa = 4.28LTYY31 pKa = 10.73SRR33 pKa = 11.84CNATVEE39 pKa = 4.86DD40 pKa = 3.5ITEE43 pKa = 4.05FLEE46 pKa = 4.3TTVASSAQCHH56 pKa = 4.98SWFAARR62 pKa = 11.84EE63 pKa = 3.92YY64 pKa = 10.52HH65 pKa = 6.49QDD67 pKa = 3.37GVPHH71 pKa = 4.9YY72 pKa = 10.37HH73 pKa = 6.96ILVKK77 pKa = 10.35FNKK80 pKa = 9.5RR81 pKa = 11.84RR82 pKa = 11.84TFRR85 pKa = 11.84NSRR88 pKa = 11.84EE89 pKa = 3.6YY90 pKa = 11.04DD91 pKa = 3.59FNGHH95 pKa = 6.04HH96 pKa = 6.83PRR98 pKa = 11.84IKK100 pKa = 10.04PVRR103 pKa = 11.84NVHH106 pKa = 7.08DD107 pKa = 3.87YY108 pKa = 10.6FRR110 pKa = 11.84YY111 pKa = 9.96ISDD114 pKa = 3.44PSKK117 pKa = 10.43PSRR120 pKa = 11.84DD121 pKa = 3.22TTFASTGWMDD131 pKa = 5.28PIPRR135 pKa = 11.84DD136 pKa = 3.33GARR139 pKa = 11.84GGGRR143 pKa = 11.84GRR145 pKa = 11.84RR146 pKa = 11.84DD147 pKa = 3.42DD148 pKa = 4.96DD149 pKa = 4.83LGGDD153 pKa = 3.55GCTWGDD159 pKa = 3.36IVRR162 pKa = 11.84ADD164 pKa = 3.61GKK166 pKa = 10.68DD167 pKa = 3.24EE168 pKa = 4.26YY169 pKa = 10.66FTLLKK174 pKa = 10.26LHH176 pKa = 6.83KK177 pKa = 10.38PRR179 pKa = 11.84DD180 pKa = 3.68YY181 pKa = 11.22CLNLQRR187 pKa = 11.84LEE189 pKa = 4.2YY190 pKa = 8.89TASRR194 pKa = 11.84LFTAITPYY202 pKa = 10.19VPEE205 pKa = 4.04YY206 pKa = 10.68EE207 pKa = 4.03YY208 pKa = 9.98DD209 pKa = 3.5TFNVPEE215 pKa = 4.74EE216 pKa = 3.94LHH218 pKa = 5.97EE219 pKa = 4.03WATVGRR225 pKa = 11.84HH226 pKa = 4.81LQPRR230 pKa = 11.84PKK232 pKa = 10.45SLIICGPSRR241 pKa = 11.84IGKK244 pKa = 6.69TEE246 pKa = 3.64WARR249 pKa = 11.84SIGLHH254 pKa = 5.32TYY256 pKa = 9.15WYY258 pKa = 8.33GQCNIDD264 pKa = 3.27SWNDD268 pKa = 2.85KK269 pKa = 10.11MEE271 pKa = 4.11YY272 pKa = 10.51LIMDD276 pKa = 4.92DD277 pKa = 5.07FSPEE281 pKa = 3.4ITKK284 pKa = 9.85YY285 pKa = 10.16LPLWKK290 pKa = 10.19GYY292 pKa = 10.49FGAQKK297 pKa = 10.09EE298 pKa = 4.36LNVTQKK304 pKa = 10.62YY305 pKa = 9.78RR306 pKa = 11.84GIYY309 pKa = 7.96TKK311 pKa = 10.71LFRR314 pKa = 11.84VPVLWLCNQIPILKK328 pKa = 9.96DD329 pKa = 3.28WEE331 pKa = 4.32QEE333 pKa = 4.09WLNMNADD340 pKa = 3.85TYY342 pKa = 10.87FINDD346 pKa = 3.45KK347 pKa = 11.11LYY349 pKa = 10.79GG350 pKa = 3.74
MM1 pKa = 7.25QNTHH5 pKa = 6.46ASSDD9 pKa = 3.91EE10 pKa = 4.05EE11 pKa = 4.63VEE13 pKa = 4.3AGPSRR18 pKa = 11.84TFRR21 pKa = 11.84IDD23 pKa = 3.11ARR25 pKa = 11.84HH26 pKa = 6.5LEE28 pKa = 4.28LTYY31 pKa = 10.73SRR33 pKa = 11.84CNATVEE39 pKa = 4.86DD40 pKa = 3.5ITEE43 pKa = 4.05FLEE46 pKa = 4.3TTVASSAQCHH56 pKa = 4.98SWFAARR62 pKa = 11.84EE63 pKa = 3.92YY64 pKa = 10.52HH65 pKa = 6.49QDD67 pKa = 3.37GVPHH71 pKa = 4.9YY72 pKa = 10.37HH73 pKa = 6.96ILVKK77 pKa = 10.35FNKK80 pKa = 9.5RR81 pKa = 11.84RR82 pKa = 11.84TFRR85 pKa = 11.84NSRR88 pKa = 11.84EE89 pKa = 3.6YY90 pKa = 11.04DD91 pKa = 3.59FNGHH95 pKa = 6.04HH96 pKa = 6.83PRR98 pKa = 11.84IKK100 pKa = 10.04PVRR103 pKa = 11.84NVHH106 pKa = 7.08DD107 pKa = 3.87YY108 pKa = 10.6FRR110 pKa = 11.84YY111 pKa = 9.96ISDD114 pKa = 3.44PSKK117 pKa = 10.43PSRR120 pKa = 11.84DD121 pKa = 3.22TTFASTGWMDD131 pKa = 5.28PIPRR135 pKa = 11.84DD136 pKa = 3.33GARR139 pKa = 11.84GGGRR143 pKa = 11.84GRR145 pKa = 11.84RR146 pKa = 11.84DD147 pKa = 3.42DD148 pKa = 4.96DD149 pKa = 4.83LGGDD153 pKa = 3.55GCTWGDD159 pKa = 3.36IVRR162 pKa = 11.84ADD164 pKa = 3.61GKK166 pKa = 10.68DD167 pKa = 3.24EE168 pKa = 4.26YY169 pKa = 10.66FTLLKK174 pKa = 10.26LHH176 pKa = 6.83KK177 pKa = 10.38PRR179 pKa = 11.84DD180 pKa = 3.68YY181 pKa = 11.22CLNLQRR187 pKa = 11.84LEE189 pKa = 4.2YY190 pKa = 8.89TASRR194 pKa = 11.84LFTAITPYY202 pKa = 10.19VPEE205 pKa = 4.04YY206 pKa = 10.68EE207 pKa = 4.03YY208 pKa = 9.98DD209 pKa = 3.5TFNVPEE215 pKa = 4.74EE216 pKa = 3.94LHH218 pKa = 5.97EE219 pKa = 4.03WATVGRR225 pKa = 11.84HH226 pKa = 4.81LQPRR230 pKa = 11.84PKK232 pKa = 10.45SLIICGPSRR241 pKa = 11.84IGKK244 pKa = 6.69TEE246 pKa = 3.64WARR249 pKa = 11.84SIGLHH254 pKa = 5.32TYY256 pKa = 9.15WYY258 pKa = 8.33GQCNIDD264 pKa = 3.27SWNDD268 pKa = 2.85KK269 pKa = 10.11MEE271 pKa = 4.11YY272 pKa = 10.51LIMDD276 pKa = 4.92DD277 pKa = 5.07FSPEE281 pKa = 3.4ITKK284 pKa = 9.85YY285 pKa = 10.16LPLWKK290 pKa = 10.19GYY292 pKa = 10.49FGAQKK297 pKa = 10.09EE298 pKa = 4.36LNVTQKK304 pKa = 10.62YY305 pKa = 9.78RR306 pKa = 11.84GIYY309 pKa = 7.96TKK311 pKa = 10.71LFRR314 pKa = 11.84VPVLWLCNQIPILKK328 pKa = 9.96DD329 pKa = 3.28WEE331 pKa = 4.32QEE333 pKa = 4.09WLNMNADD340 pKa = 3.85TYY342 pKa = 10.87FINDD346 pKa = 3.45KK347 pKa = 11.11LYY349 pKa = 10.79GG350 pKa = 3.74
Molecular weight: 41.08 kDa
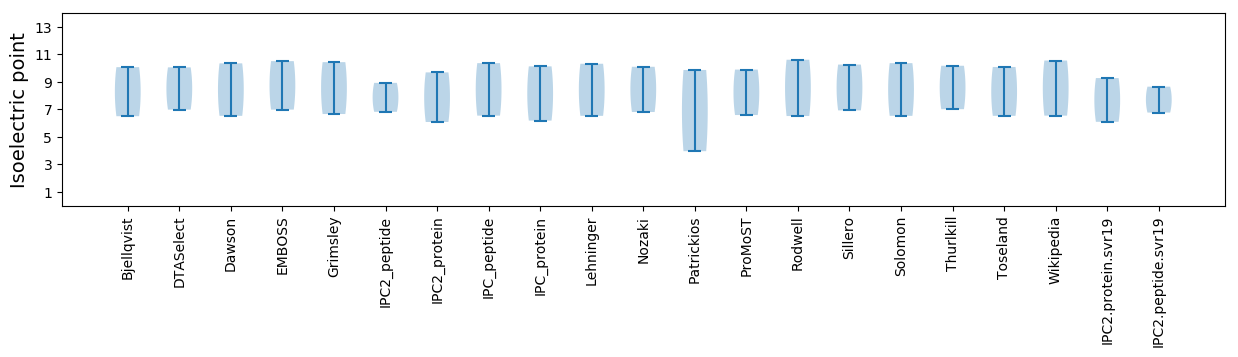
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126GA35|A0A126GA35_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-51 OX=1685781 PE=3 SV=1
MM1 pKa = 7.62PPYY4 pKa = 10.26APRR7 pKa = 11.84YY8 pKa = 7.54RR9 pKa = 11.84ASKK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VGRR19 pKa = 11.84KK20 pKa = 7.45FRR22 pKa = 11.84GRR24 pKa = 11.84KK25 pKa = 5.2RR26 pKa = 11.84TRR28 pKa = 11.84RR29 pKa = 11.84GRR31 pKa = 11.84GGQRR35 pKa = 11.84SVQRR39 pKa = 11.84YY40 pKa = 8.13SNLVRR45 pKa = 11.84LIKK48 pKa = 10.39KK49 pKa = 8.53VAIRR53 pKa = 11.84SAEE56 pKa = 4.07SKK58 pKa = 10.69FNYY61 pKa = 10.2LYY63 pKa = 10.94GSQLTALAGQGYY75 pKa = 10.5LMNPWMGITEE85 pKa = 4.3GTSQEE90 pKa = 3.81QRR92 pKa = 11.84VGNEE96 pKa = 3.94VYY98 pKa = 10.88VKK100 pKa = 10.97GMLLKK105 pKa = 11.16GNITCSANYY114 pKa = 9.51PIDD117 pKa = 3.89DD118 pKa = 3.58RR119 pKa = 11.84TVRR122 pKa = 11.84ISLIWCPDD130 pKa = 3.04NLVDD134 pKa = 4.05HH135 pKa = 6.08THH137 pKa = 5.33VSAFVRR143 pKa = 11.84YY144 pKa = 10.19DD145 pKa = 3.55NIVDD149 pKa = 3.79NEE151 pKa = 4.19GNLYY155 pKa = 9.48IDD157 pKa = 2.95NWYY160 pKa = 10.56QYY162 pKa = 10.71GVLPNSIGYY171 pKa = 10.6ANASFDD177 pKa = 4.05KK178 pKa = 10.92DD179 pKa = 3.19SGIAVLYY186 pKa = 8.62QKK188 pKa = 10.43RR189 pKa = 11.84IKK191 pKa = 10.3LRR193 pKa = 11.84QSHH196 pKa = 5.01NTWRR200 pKa = 11.84SDD202 pKa = 3.17TNLAGVSLHH211 pKa = 7.44PIRR214 pKa = 11.84CWVPMKK220 pKa = 10.36LRR222 pKa = 11.84KK223 pKa = 9.25FKK225 pKa = 10.9FVGDD229 pKa = 4.18GGNDD233 pKa = 3.24VYY235 pKa = 10.87RR236 pKa = 11.84GSKK239 pKa = 8.33GQYY242 pKa = 7.77YY243 pKa = 9.57WLFDD247 pKa = 3.46VTDD250 pKa = 3.58TFGTVASTPMLAFDD264 pKa = 5.61WISKK268 pKa = 10.25VYY270 pKa = 10.49FKK272 pKa = 11.21DD273 pKa = 3.31PP274 pKa = 3.49
MM1 pKa = 7.62PPYY4 pKa = 10.26APRR7 pKa = 11.84YY8 pKa = 7.54RR9 pKa = 11.84ASKK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VGRR19 pKa = 11.84KK20 pKa = 7.45FRR22 pKa = 11.84GRR24 pKa = 11.84KK25 pKa = 5.2RR26 pKa = 11.84TRR28 pKa = 11.84RR29 pKa = 11.84GRR31 pKa = 11.84GGQRR35 pKa = 11.84SVQRR39 pKa = 11.84YY40 pKa = 8.13SNLVRR45 pKa = 11.84LIKK48 pKa = 10.39KK49 pKa = 8.53VAIRR53 pKa = 11.84SAEE56 pKa = 4.07SKK58 pKa = 10.69FNYY61 pKa = 10.2LYY63 pKa = 10.94GSQLTALAGQGYY75 pKa = 10.5LMNPWMGITEE85 pKa = 4.3GTSQEE90 pKa = 3.81QRR92 pKa = 11.84VGNEE96 pKa = 3.94VYY98 pKa = 10.88VKK100 pKa = 10.97GMLLKK105 pKa = 11.16GNITCSANYY114 pKa = 9.51PIDD117 pKa = 3.89DD118 pKa = 3.58RR119 pKa = 11.84TVRR122 pKa = 11.84ISLIWCPDD130 pKa = 3.04NLVDD134 pKa = 4.05HH135 pKa = 6.08THH137 pKa = 5.33VSAFVRR143 pKa = 11.84YY144 pKa = 10.19DD145 pKa = 3.55NIVDD149 pKa = 3.79NEE151 pKa = 4.19GNLYY155 pKa = 9.48IDD157 pKa = 2.95NWYY160 pKa = 10.56QYY162 pKa = 10.71GVLPNSIGYY171 pKa = 10.6ANASFDD177 pKa = 4.05KK178 pKa = 10.92DD179 pKa = 3.19SGIAVLYY186 pKa = 8.62QKK188 pKa = 10.43RR189 pKa = 11.84IKK191 pKa = 10.3LRR193 pKa = 11.84QSHH196 pKa = 5.01NTWRR200 pKa = 11.84SDD202 pKa = 3.17TNLAGVSLHH211 pKa = 7.44PIRR214 pKa = 11.84CWVPMKK220 pKa = 10.36LRR222 pKa = 11.84KK223 pKa = 9.25FKK225 pKa = 10.9FVGDD229 pKa = 4.18GGNDD233 pKa = 3.24VYY235 pKa = 10.87RR236 pKa = 11.84GSKK239 pKa = 8.33GQYY242 pKa = 7.77YY243 pKa = 9.57WLFDD247 pKa = 3.46VTDD250 pKa = 3.58TFGTVASTPMLAFDD264 pKa = 5.61WISKK268 pKa = 10.25VYY270 pKa = 10.49FKK272 pKa = 11.21DD273 pKa = 3.31PP274 pKa = 3.49
Molecular weight: 31.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
624 |
274 |
350 |
312.0 |
36.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.968 ± 0.082 | 1.603 ± 0.293 |
6.731 ± 0.514 | 4.327 ± 1.443 |
4.006 ± 0.206 | 7.532 ± 0.708 |
2.724 ± 0.729 | 5.288 ± 0.103 |
5.449 ± 0.436 | 7.212 ± 0.16 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.246 | 4.968 ± 0.503 |
4.808 ± 0.457 | 3.205 ± 0.256 |
8.814 ± 0.6 | 5.929 ± 0.58 |
5.929 ± 0.684 | 5.609 ± 1.186 |
2.885 ± 0.19 | 6.25 ± 0.184 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
