
Phycisphaera sp.
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Phycisphaerae; Phycisphaerales; Phycisphaeraceae; Phycisphaera; unclassified Phycisphaera
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
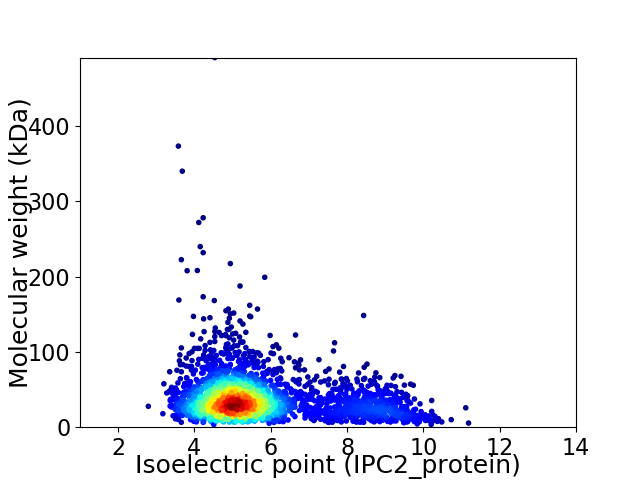
Virtual 2D-PAGE plot for 2755 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M8FWW9|A0A3M8FWW9_9BACT Uncharacterized protein OS=Phycisphaera sp. OX=2030824 GN=ED559_12255 PE=4 SV=1
MM1 pKa = 7.0VFSHH5 pKa = 6.91AAAAQDD11 pKa = 3.91VVYY14 pKa = 10.85LPVFYY19 pKa = 11.12VGTNLSADD27 pKa = 3.43GSVVVGNQVGSYY39 pKa = 7.31EE40 pKa = 4.26TFRR43 pKa = 11.84WTASSGPVLLGEE55 pKa = 4.11ATAPVLGVGAGGPDD69 pKa = 3.41VSHH72 pKa = 7.65DD73 pKa = 3.75GTRR76 pKa = 11.84VSATILTEE84 pKa = 4.27DD85 pKa = 3.84ATLATPGFWTQGLGWTRR102 pKa = 11.84CMPPMAPDD110 pKa = 4.17GRR112 pKa = 11.84ILDD115 pKa = 3.89QAIGSAWGISGDD127 pKa = 3.47GTTVSGLHH135 pKa = 6.31WYY137 pKa = 10.56SMPDD141 pKa = 3.08GSYY144 pKa = 10.64GGARR148 pKa = 11.84PFTWSASSGLIQLDD162 pKa = 3.22ITAGQDD168 pKa = 2.94GRR170 pKa = 11.84ANTTNLDD177 pKa = 3.33GSVAVGWEE185 pKa = 4.28SNAFGHH191 pKa = 5.7WQPVVWRR198 pKa = 11.84NGTRR202 pKa = 11.84MQLSEE207 pKa = 3.75NDD209 pKa = 3.33AFVGCEE215 pKa = 3.73EE216 pKa = 4.26VTSDD220 pKa = 3.08GNTVVGQSWYY230 pKa = 10.2EE231 pKa = 3.64PTLNRR236 pKa = 11.84VATVWAWNGSSYY248 pKa = 11.18DD249 pKa = 3.77QNQLGVLPGTPVDD262 pKa = 4.52FGSSVAFGVSDD273 pKa = 4.38DD274 pKa = 3.47GSMIVGVNRR283 pKa = 11.84FAVSPGGPSKK293 pKa = 11.12GFIWTAEE300 pKa = 3.89TGMVEE305 pKa = 4.18AEE307 pKa = 4.83AYY309 pKa = 10.48LADD312 pKa = 4.5LGIDD316 pKa = 3.57VSDD319 pKa = 3.82EE320 pKa = 3.73MDD322 pKa = 3.54LRR324 pKa = 11.84SVDD327 pKa = 4.36GISADD332 pKa = 3.19GSTILISGWDD342 pKa = 3.48SATQIIKK349 pKa = 10.46SAIVTLTTPCVADD362 pKa = 3.7TNGDD366 pKa = 3.84GQLTPADD373 pKa = 3.53FTAWINAFNNALPEE387 pKa = 4.44CDD389 pKa = 3.44QNADD393 pKa = 3.92GACTPTDD400 pKa = 3.73FTAWINNYY408 pKa = 9.49NAGCDD413 pKa = 3.3
MM1 pKa = 7.0VFSHH5 pKa = 6.91AAAAQDD11 pKa = 3.91VVYY14 pKa = 10.85LPVFYY19 pKa = 11.12VGTNLSADD27 pKa = 3.43GSVVVGNQVGSYY39 pKa = 7.31EE40 pKa = 4.26TFRR43 pKa = 11.84WTASSGPVLLGEE55 pKa = 4.11ATAPVLGVGAGGPDD69 pKa = 3.41VSHH72 pKa = 7.65DD73 pKa = 3.75GTRR76 pKa = 11.84VSATILTEE84 pKa = 4.27DD85 pKa = 3.84ATLATPGFWTQGLGWTRR102 pKa = 11.84CMPPMAPDD110 pKa = 4.17GRR112 pKa = 11.84ILDD115 pKa = 3.89QAIGSAWGISGDD127 pKa = 3.47GTTVSGLHH135 pKa = 6.31WYY137 pKa = 10.56SMPDD141 pKa = 3.08GSYY144 pKa = 10.64GGARR148 pKa = 11.84PFTWSASSGLIQLDD162 pKa = 3.22ITAGQDD168 pKa = 2.94GRR170 pKa = 11.84ANTTNLDD177 pKa = 3.33GSVAVGWEE185 pKa = 4.28SNAFGHH191 pKa = 5.7WQPVVWRR198 pKa = 11.84NGTRR202 pKa = 11.84MQLSEE207 pKa = 3.75NDD209 pKa = 3.33AFVGCEE215 pKa = 3.73EE216 pKa = 4.26VTSDD220 pKa = 3.08GNTVVGQSWYY230 pKa = 10.2EE231 pKa = 3.64PTLNRR236 pKa = 11.84VATVWAWNGSSYY248 pKa = 11.18DD249 pKa = 3.77QNQLGVLPGTPVDD262 pKa = 4.52FGSSVAFGVSDD273 pKa = 4.38DD274 pKa = 3.47GSMIVGVNRR283 pKa = 11.84FAVSPGGPSKK293 pKa = 11.12GFIWTAEE300 pKa = 3.89TGMVEE305 pKa = 4.18AEE307 pKa = 4.83AYY309 pKa = 10.48LADD312 pKa = 4.5LGIDD316 pKa = 3.57VSDD319 pKa = 3.82EE320 pKa = 3.73MDD322 pKa = 3.54LRR324 pKa = 11.84SVDD327 pKa = 4.36GISADD332 pKa = 3.19GSTILISGWDD342 pKa = 3.48SATQIIKK349 pKa = 10.46SAIVTLTTPCVADD362 pKa = 3.7TNGDD366 pKa = 3.84GQLTPADD373 pKa = 3.53FTAWINAFNNALPEE387 pKa = 4.44CDD389 pKa = 3.44QNADD393 pKa = 3.92GACTPTDD400 pKa = 3.73FTAWINNYY408 pKa = 9.49NAGCDD413 pKa = 3.3
Molecular weight: 43.13 kDa
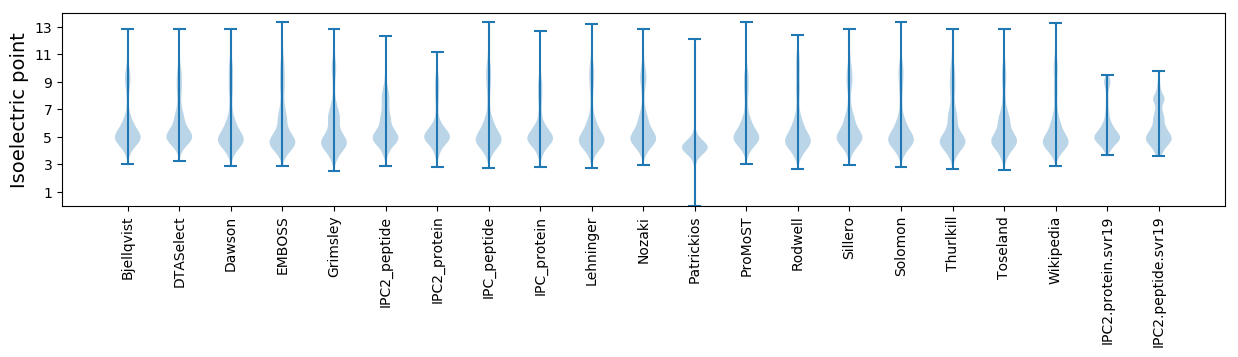
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8FWU6|A0A3M8FWU6_9BACT FHA domain-containing protein OS=Phycisphaera sp. OX=2030824 GN=ED559_11140 PE=3 SV=1
MM1 pKa = 7.08VHH3 pKa = 6.64TSPDD7 pKa = 3.04SSYY10 pKa = 11.88DD11 pKa = 3.83CLKK14 pKa = 10.3HH15 pKa = 6.13ACTRR19 pKa = 11.84RR20 pKa = 11.84PHH22 pKa = 6.46RR23 pKa = 11.84GSHH26 pKa = 4.13GRR28 pKa = 11.84IRR30 pKa = 11.84TRR32 pKa = 11.84TKK34 pKa = 9.89RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.66HH38 pKa = 4.4RR39 pKa = 11.84WQRR42 pKa = 11.84KK43 pKa = 7.29LLARR47 pKa = 11.84KK48 pKa = 8.75PRR50 pKa = 11.84GRR52 pKa = 11.84KK53 pKa = 8.23QLVRR57 pKa = 11.84RR58 pKa = 11.84LLRR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84GRR66 pKa = 11.84RR67 pKa = 11.84PLARR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84PARR76 pKa = 11.84RR77 pKa = 11.84QPARR81 pKa = 11.84RR82 pKa = 11.84QPARR86 pKa = 11.84RR87 pKa = 11.84QPARR91 pKa = 11.84RR92 pKa = 11.84PLARR96 pKa = 11.84RR97 pKa = 11.84PLARR101 pKa = 11.84RR102 pKa = 11.84PLARR106 pKa = 11.84RR107 pKa = 11.84PLARR111 pKa = 11.84RR112 pKa = 11.84PLARR116 pKa = 11.84RR117 pKa = 11.84PLARR121 pKa = 11.84RR122 pKa = 11.84PPARR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84PARR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84PARR136 pKa = 11.84RR137 pKa = 11.84PLARR141 pKa = 11.84RR142 pKa = 11.84PLARR146 pKa = 11.84RR147 pKa = 11.84RR148 pKa = 11.84PARR151 pKa = 11.84RR152 pKa = 11.84PLARR156 pKa = 11.84RR157 pKa = 11.84QPARR161 pKa = 11.84RR162 pKa = 11.84PLARR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84PARR171 pKa = 11.84RR172 pKa = 11.84PLAAAKK178 pKa = 10.34ANRR181 pKa = 11.84LSDD184 pKa = 3.99RR185 pKa = 11.84LSLSPILMLNLAAGPSGPAVFSCARR210 pKa = 11.84GRR212 pKa = 11.84ALFVRR217 pKa = 11.84DD218 pKa = 3.43
MM1 pKa = 7.08VHH3 pKa = 6.64TSPDD7 pKa = 3.04SSYY10 pKa = 11.88DD11 pKa = 3.83CLKK14 pKa = 10.3HH15 pKa = 6.13ACTRR19 pKa = 11.84RR20 pKa = 11.84PHH22 pKa = 6.46RR23 pKa = 11.84GSHH26 pKa = 4.13GRR28 pKa = 11.84IRR30 pKa = 11.84TRR32 pKa = 11.84TKK34 pKa = 9.89RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.66HH38 pKa = 4.4RR39 pKa = 11.84WQRR42 pKa = 11.84KK43 pKa = 7.29LLARR47 pKa = 11.84KK48 pKa = 8.75PRR50 pKa = 11.84GRR52 pKa = 11.84KK53 pKa = 8.23QLVRR57 pKa = 11.84RR58 pKa = 11.84LLRR61 pKa = 11.84RR62 pKa = 11.84PRR64 pKa = 11.84GRR66 pKa = 11.84RR67 pKa = 11.84PLARR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84PARR76 pKa = 11.84RR77 pKa = 11.84QPARR81 pKa = 11.84RR82 pKa = 11.84QPARR86 pKa = 11.84RR87 pKa = 11.84QPARR91 pKa = 11.84RR92 pKa = 11.84PLARR96 pKa = 11.84RR97 pKa = 11.84PLARR101 pKa = 11.84RR102 pKa = 11.84PLARR106 pKa = 11.84RR107 pKa = 11.84PLARR111 pKa = 11.84RR112 pKa = 11.84PLARR116 pKa = 11.84RR117 pKa = 11.84PLARR121 pKa = 11.84RR122 pKa = 11.84PPARR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84PARR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84PARR136 pKa = 11.84RR137 pKa = 11.84PLARR141 pKa = 11.84RR142 pKa = 11.84PLARR146 pKa = 11.84RR147 pKa = 11.84RR148 pKa = 11.84PARR151 pKa = 11.84RR152 pKa = 11.84PLARR156 pKa = 11.84RR157 pKa = 11.84QPARR161 pKa = 11.84RR162 pKa = 11.84PLARR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84PARR171 pKa = 11.84RR172 pKa = 11.84PLAAAKK178 pKa = 10.34ANRR181 pKa = 11.84LSDD184 pKa = 3.99RR185 pKa = 11.84LSLSPILMLNLAAGPSGPAVFSCARR210 pKa = 11.84GRR212 pKa = 11.84ALFVRR217 pKa = 11.84DD218 pKa = 3.43
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989657 |
31 |
4552 |
359.2 |
39.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.218 ± 0.053 | 1.033 ± 0.016 |
6.561 ± 0.055 | 6.654 ± 0.053 |
3.496 ± 0.03 | 8.445 ± 0.054 |
2.044 ± 0.029 | 5.506 ± 0.033 |
3.263 ± 0.044 | 9.341 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.027 | 3.081 ± 0.039 |
4.948 ± 0.033 | 3.279 ± 0.027 |
6.54 ± 0.046 | 6.522 ± 0.038 |
5.749 ± 0.043 | 7.395 ± 0.035 |
1.326 ± 0.022 | 2.241 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
