
Citrus chlorotic dwarf associated virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Citlodavirus
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
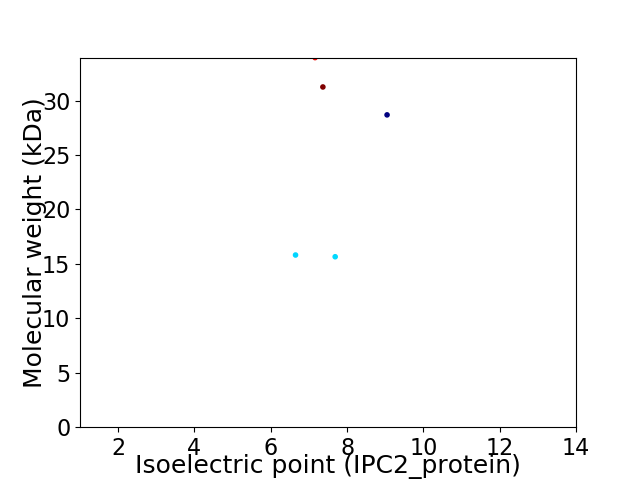
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6X8D9|I6X8D9_9GEMI Replication-associated protein (Fragment) OS=Citrus chlorotic dwarf associated virus OX=1202142 PE=4 SV=1
MM1 pKa = 7.37CHH3 pKa = 6.11YY4 pKa = 10.41ALSVQDD10 pKa = 4.22LPEE13 pKa = 4.26SLFGLMSMLSVRR25 pKa = 11.84YY26 pKa = 8.35LKK28 pKa = 10.63CVEE31 pKa = 3.93EE32 pKa = 5.02RR33 pKa = 11.84EE34 pKa = 4.35MEE36 pKa = 4.14RR37 pKa = 11.84SLMVGNPVGGALGNARR53 pKa = 11.84ILIRR57 pKa = 11.84LIRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.47CRR64 pKa = 11.84CRR66 pKa = 11.84DD67 pKa = 3.14WVRR70 pKa = 11.84KK71 pKa = 9.55GRR73 pKa = 11.84VNGEE77 pKa = 3.79YY78 pKa = 10.38QEE80 pKa = 4.47WRR82 pKa = 11.84TIWDD86 pKa = 4.3KK87 pKa = 11.28PCNDD91 pKa = 4.68CGDD94 pKa = 4.08AADD97 pKa = 4.27VGHH100 pKa = 6.76KK101 pKa = 9.21EE102 pKa = 3.93KK103 pKa = 11.08KK104 pKa = 8.14EE105 pKa = 3.78AQVAEE110 pKa = 4.42KK111 pKa = 10.63GKK113 pKa = 9.84EE114 pKa = 3.94AQDD117 pKa = 3.4CWGCICEE124 pKa = 5.02GPAQEE129 pKa = 5.4KK130 pKa = 10.29VEE132 pKa = 4.08QRR134 pKa = 11.84CSSGVV139 pKa = 3.12
MM1 pKa = 7.37CHH3 pKa = 6.11YY4 pKa = 10.41ALSVQDD10 pKa = 4.22LPEE13 pKa = 4.26SLFGLMSMLSVRR25 pKa = 11.84YY26 pKa = 8.35LKK28 pKa = 10.63CVEE31 pKa = 3.93EE32 pKa = 5.02RR33 pKa = 11.84EE34 pKa = 4.35MEE36 pKa = 4.14RR37 pKa = 11.84SLMVGNPVGGALGNARR53 pKa = 11.84ILIRR57 pKa = 11.84LIRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.47CRR64 pKa = 11.84CRR66 pKa = 11.84DD67 pKa = 3.14WVRR70 pKa = 11.84KK71 pKa = 9.55GRR73 pKa = 11.84VNGEE77 pKa = 3.79YY78 pKa = 10.38QEE80 pKa = 4.47WRR82 pKa = 11.84TIWDD86 pKa = 4.3KK87 pKa = 11.28PCNDD91 pKa = 4.68CGDD94 pKa = 4.08AADD97 pKa = 4.27VGHH100 pKa = 6.76KK101 pKa = 9.21EE102 pKa = 3.93KK103 pKa = 11.08KK104 pKa = 8.14EE105 pKa = 3.78AQVAEE110 pKa = 4.42KK111 pKa = 10.63GKK113 pKa = 9.84EE114 pKa = 3.94AQDD117 pKa = 3.4CWGCICEE124 pKa = 5.02GPAQEE129 pKa = 5.4KK130 pKa = 10.29VEE132 pKa = 4.08QRR134 pKa = 11.84CSSGVV139 pKa = 3.12
Molecular weight: 15.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6XMA9|I6XMA9_9GEMI Movement protein BC1 OS=Citrus chlorotic dwarf associated virus OX=1202142 PE=3 SV=1
MM1 pKa = 7.27VSTRR5 pKa = 11.84SGGQYY10 pKa = 7.93GTSRR14 pKa = 11.84VTTVEE19 pKa = 4.29TLPMWATKK27 pKa = 9.65RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84WPRR36 pKa = 11.84KK37 pKa = 7.42EE38 pKa = 4.19KK39 pKa = 10.34KK40 pKa = 9.58PKK42 pKa = 8.88IAGAVFVKK50 pKa = 10.33ARR52 pKa = 11.84PRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.35SSKK59 pKa = 10.14GVPPGCKK66 pKa = 9.88GPCKK70 pKa = 9.45THH72 pKa = 5.31TVDD75 pKa = 3.77VIKK78 pKa = 9.89TIYY81 pKa = 10.0HH82 pKa = 7.04DD83 pKa = 3.92GRR85 pKa = 11.84GSGMISNIDD94 pKa = 3.42RR95 pKa = 11.84GDD97 pKa = 3.52EE98 pKa = 3.84LGQRR102 pKa = 11.84EE103 pKa = 4.13GRR105 pKa = 11.84KK106 pKa = 9.19IRR108 pKa = 11.84VSRR111 pKa = 11.84MIIRR115 pKa = 11.84GKK117 pKa = 9.83IWLDD121 pKa = 3.25VNNASVPGSNLAKK134 pKa = 10.0IWIFKK139 pKa = 9.87DD140 pKa = 3.25RR141 pKa = 11.84RR142 pKa = 11.84PGTEE146 pKa = 3.16PVAFNALMDD155 pKa = 4.0MSDD158 pKa = 4.42SEE160 pKa = 4.32PLSAFVKK167 pKa = 9.91VDD169 pKa = 3.05YY170 pKa = 10.25RR171 pKa = 11.84DD172 pKa = 3.29RR173 pKa = 11.84FIALHH178 pKa = 5.79TMTVDD183 pKa = 3.0LHH185 pKa = 6.18GGKK188 pKa = 10.07DD189 pKa = 3.64FRR191 pKa = 11.84VDD193 pKa = 3.82EE194 pKa = 4.86LDD196 pKa = 3.57LDD198 pKa = 4.05EE199 pKa = 5.33LVEE202 pKa = 4.28INSDD206 pKa = 3.64VLFSHH211 pKa = 7.09EE212 pKa = 4.67DD213 pKa = 3.56DD214 pKa = 4.3GSVAHH219 pKa = 7.03TIQNGIFIYY228 pKa = 8.78YY229 pKa = 9.6ACSDD233 pKa = 3.54PRR235 pKa = 11.84QTVQITAQARR245 pKa = 11.84LYY247 pKa = 10.75FYY249 pKa = 10.96DD250 pKa = 3.54STSNN254 pKa = 3.23
MM1 pKa = 7.27VSTRR5 pKa = 11.84SGGQYY10 pKa = 7.93GTSRR14 pKa = 11.84VTTVEE19 pKa = 4.29TLPMWATKK27 pKa = 9.65RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84PRR33 pKa = 11.84WPRR36 pKa = 11.84KK37 pKa = 7.42EE38 pKa = 4.19KK39 pKa = 10.34KK40 pKa = 9.58PKK42 pKa = 8.88IAGAVFVKK50 pKa = 10.33ARR52 pKa = 11.84PRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 9.35SSKK59 pKa = 10.14GVPPGCKK66 pKa = 9.88GPCKK70 pKa = 9.45THH72 pKa = 5.31TVDD75 pKa = 3.77VIKK78 pKa = 9.89TIYY81 pKa = 10.0HH82 pKa = 7.04DD83 pKa = 3.92GRR85 pKa = 11.84GSGMISNIDD94 pKa = 3.42RR95 pKa = 11.84GDD97 pKa = 3.52EE98 pKa = 3.84LGQRR102 pKa = 11.84EE103 pKa = 4.13GRR105 pKa = 11.84KK106 pKa = 9.19IRR108 pKa = 11.84VSRR111 pKa = 11.84MIIRR115 pKa = 11.84GKK117 pKa = 9.83IWLDD121 pKa = 3.25VNNASVPGSNLAKK134 pKa = 10.0IWIFKK139 pKa = 9.87DD140 pKa = 3.25RR141 pKa = 11.84RR142 pKa = 11.84PGTEE146 pKa = 3.16PVAFNALMDD155 pKa = 4.0MSDD158 pKa = 4.42SEE160 pKa = 4.32PLSAFVKK167 pKa = 9.91VDD169 pKa = 3.05YY170 pKa = 10.25RR171 pKa = 11.84DD172 pKa = 3.29RR173 pKa = 11.84FIALHH178 pKa = 5.79TMTVDD183 pKa = 3.0LHH185 pKa = 6.18GGKK188 pKa = 10.07DD189 pKa = 3.64FRR191 pKa = 11.84VDD193 pKa = 3.82EE194 pKa = 4.86LDD196 pKa = 3.57LDD198 pKa = 4.05EE199 pKa = 5.33LVEE202 pKa = 4.28INSDD206 pKa = 3.64VLFSHH211 pKa = 7.09EE212 pKa = 4.67DD213 pKa = 3.56DD214 pKa = 4.3GSVAHH219 pKa = 7.03TIQNGIFIYY228 pKa = 8.78YY229 pKa = 9.6ACSDD233 pKa = 3.54PRR235 pKa = 11.84QTVQITAQARR245 pKa = 11.84LYY247 pKa = 10.75FYY249 pKa = 10.96DD250 pKa = 3.54STSNN254 pKa = 3.23
Molecular weight: 28.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103 |
135 |
306 |
220.6 |
25.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.074 ± 0.44 | 3.173 ± 0.867 |
6.346 ± 0.774 | 5.621 ± 1.078 |
3.626 ± 0.737 | 5.984 ± 1.082 |
2.539 ± 0.327 | 5.44 ± 0.767 |
6.346 ± 0.288 | 6.981 ± 0.65 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.306 | 4.17 ± 0.448 |
5.802 ± 0.745 | 3.717 ± 0.408 |
7.525 ± 0.869 | 8.432 ± 1.075 |
3.989 ± 0.891 | 6.437 ± 0.766 |
1.813 ± 0.265 | 3.445 ± 0.528 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
