
bacterium HR38
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
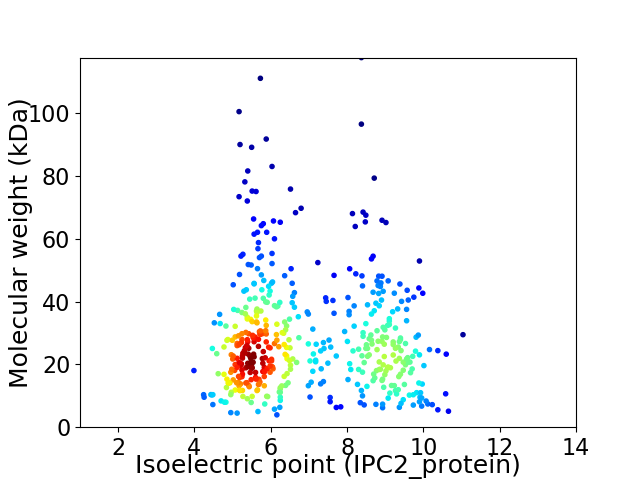
Virtual 2D-PAGE plot for 485 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
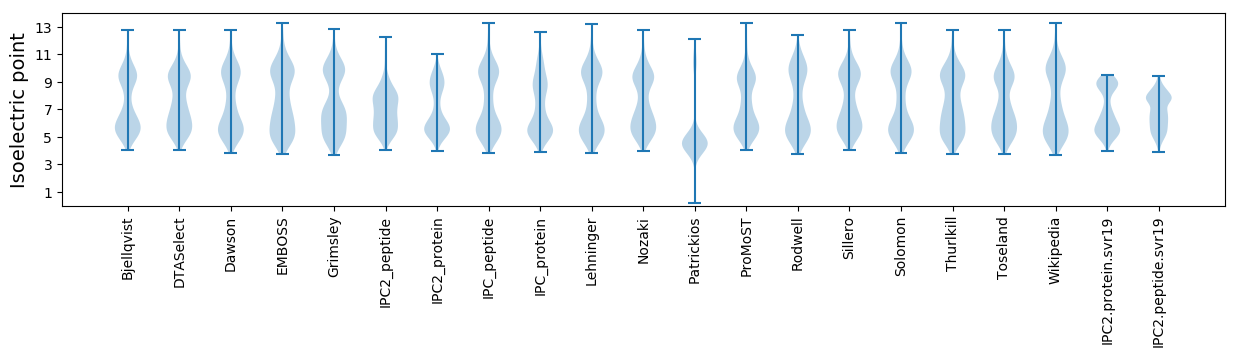
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6AQR4|A0A2H6AQR4_9BACT Antitoxin VapB41 OS=bacterium HR38 OX=2035433 GN=HRbin38_00073 PE=4 SV=1
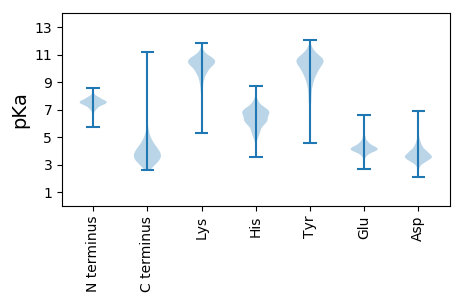
MM1 pKa = 8.22DD2 pKa = 4.5YY3 pKa = 11.38SEE5 pKa = 4.93MPYY8 pKa = 10.3QEE10 pKa = 4.3ARR12 pKa = 11.84KK13 pKa = 8.94QAVKK17 pKa = 10.7VLEE20 pKa = 4.81DD21 pKa = 4.13GYY23 pKa = 11.53GDD25 pKa = 4.38AVILKK30 pKa = 9.4DD31 pKa = 3.56AHH33 pKa = 6.71GYY35 pKa = 6.16WALYY39 pKa = 8.94YY40 pKa = 10.52LYY42 pKa = 9.88GFQVPPPEE50 pKa = 5.1APPHH54 pKa = 5.16WMEE57 pKa = 4.33GPFPGEE63 pKa = 3.53EE64 pKa = 4.79GIRR67 pKa = 11.84SPYY70 pKa = 10.42EE71 pKa = 3.46MQKK74 pKa = 10.54FLEE77 pKa = 4.31EE78 pKa = 4.15QGDD81 pKa = 3.86FTYY84 pKa = 11.16LNDD87 pKa = 3.21VDD89 pKa = 4.15
MM1 pKa = 8.22DD2 pKa = 4.5YY3 pKa = 11.38SEE5 pKa = 4.93MPYY8 pKa = 10.3QEE10 pKa = 4.3ARR12 pKa = 11.84KK13 pKa = 8.94QAVKK17 pKa = 10.7VLEE20 pKa = 4.81DD21 pKa = 4.13GYY23 pKa = 11.53GDD25 pKa = 4.38AVILKK30 pKa = 9.4DD31 pKa = 3.56AHH33 pKa = 6.71GYY35 pKa = 6.16WALYY39 pKa = 8.94YY40 pKa = 10.52LYY42 pKa = 9.88GFQVPPPEE50 pKa = 5.1APPHH54 pKa = 5.16WMEE57 pKa = 4.33GPFPGEE63 pKa = 3.53EE64 pKa = 4.79GIRR67 pKa = 11.84SPYY70 pKa = 10.42EE71 pKa = 3.46MQKK74 pKa = 10.54FLEE77 pKa = 4.31EE78 pKa = 4.15QGDD81 pKa = 3.86FTYY84 pKa = 11.16LNDD87 pKa = 3.21VDD89 pKa = 4.15
Molecular weight: 10.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6ARN6|A0A2H6ARN6_9BACT Ribose import ATP-binding protein RbsA OS=bacterium HR38 OX=2035433 GN=rbsA PE=4 SV=1
MM1 pKa = 7.67ARR3 pKa = 11.84RR4 pKa = 11.84ALWGSGPTTFCTSMRR19 pKa = 11.84FSVRR23 pKa = 11.84VPVLSVAITVTLPKK37 pKa = 10.45ASTAARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84TTALRR50 pKa = 11.84RR51 pKa = 11.84AMRR54 pKa = 11.84STPRR58 pKa = 11.84AKK60 pKa = 10.61ARR62 pKa = 11.84VSTAGSPSGTAATARR77 pKa = 11.84ATAKK81 pKa = 10.07RR82 pKa = 11.84RR83 pKa = 11.84TWAEE87 pKa = 3.76KK88 pKa = 9.7PSARR92 pKa = 11.84KK93 pKa = 9.09PSATRR98 pKa = 11.84AAASPITQRR107 pKa = 11.84ATLWPKK113 pKa = 9.1PSRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84SRR120 pKa = 11.84GVFSSRR126 pKa = 11.84IPAKK130 pKa = 10.21SPAMRR135 pKa = 11.84PMALLRR141 pKa = 11.84PVRR144 pKa = 11.84LTSIRR149 pKa = 11.84ALPRR153 pKa = 11.84TNRR156 pKa = 11.84VPEE159 pKa = 4.28KK160 pKa = 9.77TSSPTLFSTGTLSPVRR176 pKa = 11.84RR177 pKa = 11.84LSSTKK182 pKa = 9.72TPLASRR188 pKa = 11.84STPSAGMRR196 pKa = 11.84SPASRR201 pKa = 11.84RR202 pKa = 11.84TTSPGTISRR211 pKa = 11.84LGRR214 pKa = 11.84CWKK217 pKa = 10.01RR218 pKa = 11.84PSRR221 pKa = 11.84RR222 pKa = 11.84TQAMGSARR230 pKa = 11.84ALRR233 pKa = 11.84RR234 pKa = 11.84LRR236 pKa = 11.84ASSEE240 pKa = 3.99RR241 pKa = 11.84FSCRR245 pKa = 11.84VPRR248 pKa = 11.84VALRR252 pKa = 11.84IRR254 pKa = 11.84MARR257 pKa = 11.84MAKK260 pKa = 10.21ASRR263 pKa = 11.84GSPWAPSS270 pKa = 3.14
MM1 pKa = 7.67ARR3 pKa = 11.84RR4 pKa = 11.84ALWGSGPTTFCTSMRR19 pKa = 11.84FSVRR23 pKa = 11.84VPVLSVAITVTLPKK37 pKa = 10.45ASTAARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84TTALRR50 pKa = 11.84RR51 pKa = 11.84AMRR54 pKa = 11.84STPRR58 pKa = 11.84AKK60 pKa = 10.61ARR62 pKa = 11.84VSTAGSPSGTAATARR77 pKa = 11.84ATAKK81 pKa = 10.07RR82 pKa = 11.84RR83 pKa = 11.84TWAEE87 pKa = 3.76KK88 pKa = 9.7PSARR92 pKa = 11.84KK93 pKa = 9.09PSATRR98 pKa = 11.84AAASPITQRR107 pKa = 11.84ATLWPKK113 pKa = 9.1PSRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84SRR120 pKa = 11.84GVFSSRR126 pKa = 11.84IPAKK130 pKa = 10.21SPAMRR135 pKa = 11.84PMALLRR141 pKa = 11.84PVRR144 pKa = 11.84LTSIRR149 pKa = 11.84ALPRR153 pKa = 11.84TNRR156 pKa = 11.84VPEE159 pKa = 4.28KK160 pKa = 9.77TSSPTLFSTGTLSPVRR176 pKa = 11.84RR177 pKa = 11.84LSSTKK182 pKa = 9.72TPLASRR188 pKa = 11.84STPSAGMRR196 pKa = 11.84SPASRR201 pKa = 11.84RR202 pKa = 11.84TTSPGTISRR211 pKa = 11.84LGRR214 pKa = 11.84CWKK217 pKa = 10.01RR218 pKa = 11.84PSRR221 pKa = 11.84RR222 pKa = 11.84TQAMGSARR230 pKa = 11.84ALRR233 pKa = 11.84RR234 pKa = 11.84LRR236 pKa = 11.84ASSEE240 pKa = 3.99RR241 pKa = 11.84FSCRR245 pKa = 11.84VPRR248 pKa = 11.84VALRR252 pKa = 11.84IRR254 pKa = 11.84MARR257 pKa = 11.84MAKK260 pKa = 10.21ASRR263 pKa = 11.84GSPWAPSS270 pKa = 3.14
Molecular weight: 29.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
126724 |
34 |
1061 |
261.3 |
28.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.603 ± 0.123 | 0.485 ± 0.036 |
3.507 ± 0.068 | 8.219 ± 0.14 |
3.946 ± 0.072 | 9.546 ± 0.138 |
2.053 ± 0.061 | 2.942 ± 0.093 |
3.658 ± 0.083 | 14.853 ± 0.215 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.663 ± 0.039 | 1.605 ± 0.06 |
6.474 ± 0.101 | 3.051 ± 0.082 |
7.752 ± 0.126 | 3.89 ± 0.07 |
3.786 ± 0.08 | 7.662 ± 0.114 |
1.478 ± 0.059 | 2.828 ± 0.058 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
