
Hubei narna-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
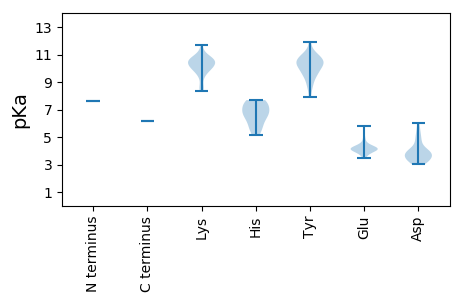
Protein with the lowest isoelectric point:
>tr|A0A1L3KIU3|A0A1L3KIU3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 4 OX=1922958 PE=4 SV=1
MM1 pKa = 7.64FIFEE5 pKa = 4.64TFSGQCKK12 pKa = 8.64EE13 pKa = 4.22WQNLCHH19 pKa = 6.45SRR21 pKa = 11.84VKK23 pKa = 10.17EE24 pKa = 3.66ALRR27 pKa = 11.84IRR29 pKa = 11.84KK30 pKa = 8.62LPACWACHH38 pKa = 5.88FPSDD42 pKa = 5.2DD43 pKa = 3.59NPEE46 pKa = 4.02YY47 pKa = 10.56LQVVTSLFNSKK58 pKa = 9.95VEE60 pKa = 4.1EE61 pKa = 4.33KK62 pKa = 10.42DD63 pKa = 3.02WPKK66 pKa = 10.52YY67 pKa = 8.74IQWAIDD73 pKa = 3.47FSTRR77 pKa = 11.84LLEE80 pKa = 4.44SYY82 pKa = 8.96LTYY85 pKa = 10.74FEE87 pKa = 5.78LGLNRR92 pKa = 11.84VLPTDD97 pKa = 3.68RR98 pKa = 11.84KK99 pKa = 9.82ILRR102 pKa = 11.84KK103 pKa = 10.23VIFDD107 pKa = 3.94CLDD110 pKa = 3.11YY111 pKa = 10.99WFLLSLIGKK120 pKa = 8.46LPKK123 pKa = 9.57YY124 pKa = 10.18LKK126 pKa = 10.47YY127 pKa = 7.9WTCKK131 pKa = 9.58FFSDD135 pKa = 4.7ILQQFDD141 pKa = 3.61QPEE144 pKa = 4.22KK145 pKa = 10.89PEE147 pKa = 4.26IPLKK151 pKa = 10.42FKK153 pKa = 10.78PFICGQIAGKK163 pKa = 9.22IHH165 pKa = 7.44LITYY169 pKa = 8.64VRR171 pKa = 11.84TRR173 pKa = 11.84SDD175 pKa = 3.26RR176 pKa = 11.84KK177 pKa = 9.93PSEE180 pKa = 3.77DD181 pKa = 3.29HH182 pKa = 6.64IKK184 pKa = 10.73FFFGLLDD191 pKa = 3.71LKK193 pKa = 10.75KK194 pKa = 10.53ASRR197 pKa = 11.84PIHH200 pKa = 6.16KK201 pKa = 9.92IFVTEE206 pKa = 3.96EE207 pKa = 3.91VEE209 pKa = 4.43KK210 pKa = 10.95SIEE213 pKa = 4.01RR214 pKa = 11.84LTTSPEE220 pKa = 3.86RR221 pKa = 11.84EE222 pKa = 3.9PLRR225 pKa = 11.84NGIIRR230 pKa = 11.84SIKK233 pKa = 9.61RR234 pKa = 11.84VCNSLVFSHH243 pKa = 6.86IRR245 pKa = 11.84GNCSDD250 pKa = 3.86IPINLPSCLPTTGGLEE266 pKa = 3.91NPRR269 pKa = 11.84RR270 pKa = 11.84TGGCYY275 pKa = 10.42GFLQGNLSSLCNNISGSFHH294 pKa = 6.77LCNMEE299 pKa = 3.92KK300 pKa = 10.39DD301 pKa = 4.0GEE303 pKa = 4.2ILPVSVPFNPRR314 pKa = 11.84IEE316 pKa = 4.05LSYY319 pKa = 10.78YY320 pKa = 10.26RR321 pKa = 11.84HH322 pKa = 6.83LDD324 pKa = 3.9DD325 pKa = 5.99LSKK328 pKa = 9.66TYY330 pKa = 10.1PGSIPVEE337 pKa = 4.06MVGLPEE343 pKa = 4.12PFKK346 pKa = 11.32VRR348 pKa = 11.84TISKK352 pKa = 10.7GPIDD356 pKa = 5.32LYY358 pKa = 11.01AQARR362 pKa = 11.84RR363 pKa = 11.84YY364 pKa = 9.71QGLFWHH370 pKa = 7.31LLRR373 pKa = 11.84HH374 pKa = 5.66SPVFSLTWSPLCEE387 pKa = 3.6YY388 pKa = 10.23HH389 pKa = 7.34LRR391 pKa = 11.84YY392 pKa = 9.83LYY394 pKa = 10.77NGNPFSKK401 pKa = 10.49NRR403 pKa = 11.84LEE405 pKa = 4.49SYY407 pKa = 10.43LLSADD412 pKa = 3.72YY413 pKa = 10.42EE414 pKa = 4.21AATDD418 pKa = 4.02SIYY421 pKa = 11.09KK422 pKa = 9.96EE423 pKa = 4.26IIEE426 pKa = 4.28EE427 pKa = 4.12TAEE430 pKa = 4.16HH431 pKa = 6.98LFHH434 pKa = 7.68LLDD437 pKa = 4.31IPWSDD442 pKa = 3.2RR443 pKa = 11.84ALLLKK448 pKa = 10.28ILTRR452 pKa = 11.84CEE454 pKa = 3.48IEE456 pKa = 4.21RR457 pKa = 11.84LDD459 pKa = 4.94GEE461 pKa = 4.38SLKK464 pKa = 10.61QKK466 pKa = 10.17RR467 pKa = 11.84GQLMGSPLSFVLLCIINAGVNLFVIEE493 pKa = 4.14KK494 pKa = 10.5SIGEE498 pKa = 4.15RR499 pKa = 11.84IKK501 pKa = 10.82FPLFGEE507 pKa = 4.23EE508 pKa = 4.83VPFLVNGDD516 pKa = 4.53DD517 pKa = 3.92YY518 pKa = 11.94LSFLPVRR525 pKa = 11.84YY526 pKa = 9.6YY527 pKa = 10.25EE528 pKa = 4.02FWKK531 pKa = 10.77EE532 pKa = 4.03CVTSVGLKK540 pKa = 10.06PSLGKK545 pKa = 10.42NYY547 pKa = 9.58LHH549 pKa = 7.57PYY551 pKa = 9.49LGTVNSTLIKK561 pKa = 10.12INRR564 pKa = 11.84PLHH567 pKa = 5.13FDD569 pKa = 3.38EE570 pKa = 5.4RR571 pKa = 11.84FKK573 pKa = 11.71DD574 pKa = 3.7NLPGKK579 pKa = 9.94IFRR582 pKa = 11.84HH583 pKa = 5.54LKK585 pKa = 10.48LSLAYY590 pKa = 9.37PPEE593 pKa = 4.27EE594 pKa = 3.6QDD596 pKa = 4.77VLIKK600 pKa = 10.78DD601 pKa = 3.9DD602 pKa = 3.64TFDD605 pKa = 3.18WRR607 pKa = 11.84KK608 pKa = 10.08ISIKK612 pKa = 10.01TNLEE616 pKa = 3.76RR617 pKa = 11.84LILDD621 pKa = 3.26QKK623 pKa = 11.33EE624 pKa = 4.2DD625 pKa = 3.35IQRR628 pKa = 11.84TLIFYY633 pKa = 10.53ALRR636 pKa = 11.84KK637 pKa = 8.86FKK639 pKa = 10.77KK640 pKa = 9.3FLSVIPKK647 pKa = 8.32EE648 pKa = 4.08IPYY651 pKa = 10.27SVSPSLGGLGIPLVDD666 pKa = 4.01HH667 pKa = 6.47KK668 pKa = 11.08QVSDD672 pKa = 3.46PHH674 pKa = 7.63RR675 pKa = 11.84MRR677 pKa = 11.84VSYY680 pKa = 10.82LITRR684 pKa = 11.84PPKK687 pKa = 10.1RR688 pKa = 11.84GRR690 pKa = 11.84TLIFPGEE697 pKa = 4.2VSQVDD702 pKa = 3.97LLLKK706 pKa = 10.4EE707 pKa = 4.88KK708 pKa = 10.54EE709 pKa = 4.16NQLLDD714 pKa = 3.5QLEE717 pKa = 4.55IPWEE721 pKa = 4.42PGTEE725 pKa = 3.99KK726 pKa = 10.58PSPRR730 pKa = 11.84EE731 pKa = 3.82GEE733 pKa = 4.37SPFSRR738 pKa = 11.84WCFLYY743 pKa = 10.22EE744 pKa = 4.07FEE746 pKa = 5.09KK747 pKa = 10.66IKK749 pKa = 10.7PKK751 pKa = 10.77VSILQTLKK759 pKa = 9.63KK760 pKa = 8.33WSYY763 pKa = 9.61IIKK766 pKa = 10.28RR767 pKa = 11.84EE768 pKa = 4.02LKK770 pKa = 10.09EE771 pKa = 4.17SNRR774 pKa = 11.84LLKK777 pKa = 10.34VLRR780 pKa = 11.84PASSTSCKK788 pKa = 9.89QDD790 pKa = 3.13VKK792 pKa = 10.3PWTRR796 pKa = 11.84TFPLYY801 pKa = 10.62TFDD804 pKa = 5.62LGLGKK809 pKa = 10.25DD810 pKa = 3.91DD811 pKa = 4.08FVSRR815 pKa = 11.84AALFRR820 pKa = 11.84EE821 pKa = 4.09ITPGIDD827 pKa = 3.55LFII830 pKa = 6.14
MM1 pKa = 7.64FIFEE5 pKa = 4.64TFSGQCKK12 pKa = 8.64EE13 pKa = 4.22WQNLCHH19 pKa = 6.45SRR21 pKa = 11.84VKK23 pKa = 10.17EE24 pKa = 3.66ALRR27 pKa = 11.84IRR29 pKa = 11.84KK30 pKa = 8.62LPACWACHH38 pKa = 5.88FPSDD42 pKa = 5.2DD43 pKa = 3.59NPEE46 pKa = 4.02YY47 pKa = 10.56LQVVTSLFNSKK58 pKa = 9.95VEE60 pKa = 4.1EE61 pKa = 4.33KK62 pKa = 10.42DD63 pKa = 3.02WPKK66 pKa = 10.52YY67 pKa = 8.74IQWAIDD73 pKa = 3.47FSTRR77 pKa = 11.84LLEE80 pKa = 4.44SYY82 pKa = 8.96LTYY85 pKa = 10.74FEE87 pKa = 5.78LGLNRR92 pKa = 11.84VLPTDD97 pKa = 3.68RR98 pKa = 11.84KK99 pKa = 9.82ILRR102 pKa = 11.84KK103 pKa = 10.23VIFDD107 pKa = 3.94CLDD110 pKa = 3.11YY111 pKa = 10.99WFLLSLIGKK120 pKa = 8.46LPKK123 pKa = 9.57YY124 pKa = 10.18LKK126 pKa = 10.47YY127 pKa = 7.9WTCKK131 pKa = 9.58FFSDD135 pKa = 4.7ILQQFDD141 pKa = 3.61QPEE144 pKa = 4.22KK145 pKa = 10.89PEE147 pKa = 4.26IPLKK151 pKa = 10.42FKK153 pKa = 10.78PFICGQIAGKK163 pKa = 9.22IHH165 pKa = 7.44LITYY169 pKa = 8.64VRR171 pKa = 11.84TRR173 pKa = 11.84SDD175 pKa = 3.26RR176 pKa = 11.84KK177 pKa = 9.93PSEE180 pKa = 3.77DD181 pKa = 3.29HH182 pKa = 6.64IKK184 pKa = 10.73FFFGLLDD191 pKa = 3.71LKK193 pKa = 10.75KK194 pKa = 10.53ASRR197 pKa = 11.84PIHH200 pKa = 6.16KK201 pKa = 9.92IFVTEE206 pKa = 3.96EE207 pKa = 3.91VEE209 pKa = 4.43KK210 pKa = 10.95SIEE213 pKa = 4.01RR214 pKa = 11.84LTTSPEE220 pKa = 3.86RR221 pKa = 11.84EE222 pKa = 3.9PLRR225 pKa = 11.84NGIIRR230 pKa = 11.84SIKK233 pKa = 9.61RR234 pKa = 11.84VCNSLVFSHH243 pKa = 6.86IRR245 pKa = 11.84GNCSDD250 pKa = 3.86IPINLPSCLPTTGGLEE266 pKa = 3.91NPRR269 pKa = 11.84RR270 pKa = 11.84TGGCYY275 pKa = 10.42GFLQGNLSSLCNNISGSFHH294 pKa = 6.77LCNMEE299 pKa = 3.92KK300 pKa = 10.39DD301 pKa = 4.0GEE303 pKa = 4.2ILPVSVPFNPRR314 pKa = 11.84IEE316 pKa = 4.05LSYY319 pKa = 10.78YY320 pKa = 10.26RR321 pKa = 11.84HH322 pKa = 6.83LDD324 pKa = 3.9DD325 pKa = 5.99LSKK328 pKa = 9.66TYY330 pKa = 10.1PGSIPVEE337 pKa = 4.06MVGLPEE343 pKa = 4.12PFKK346 pKa = 11.32VRR348 pKa = 11.84TISKK352 pKa = 10.7GPIDD356 pKa = 5.32LYY358 pKa = 11.01AQARR362 pKa = 11.84RR363 pKa = 11.84YY364 pKa = 9.71QGLFWHH370 pKa = 7.31LLRR373 pKa = 11.84HH374 pKa = 5.66SPVFSLTWSPLCEE387 pKa = 3.6YY388 pKa = 10.23HH389 pKa = 7.34LRR391 pKa = 11.84YY392 pKa = 9.83LYY394 pKa = 10.77NGNPFSKK401 pKa = 10.49NRR403 pKa = 11.84LEE405 pKa = 4.49SYY407 pKa = 10.43LLSADD412 pKa = 3.72YY413 pKa = 10.42EE414 pKa = 4.21AATDD418 pKa = 4.02SIYY421 pKa = 11.09KK422 pKa = 9.96EE423 pKa = 4.26IIEE426 pKa = 4.28EE427 pKa = 4.12TAEE430 pKa = 4.16HH431 pKa = 6.98LFHH434 pKa = 7.68LLDD437 pKa = 4.31IPWSDD442 pKa = 3.2RR443 pKa = 11.84ALLLKK448 pKa = 10.28ILTRR452 pKa = 11.84CEE454 pKa = 3.48IEE456 pKa = 4.21RR457 pKa = 11.84LDD459 pKa = 4.94GEE461 pKa = 4.38SLKK464 pKa = 10.61QKK466 pKa = 10.17RR467 pKa = 11.84GQLMGSPLSFVLLCIINAGVNLFVIEE493 pKa = 4.14KK494 pKa = 10.5SIGEE498 pKa = 4.15RR499 pKa = 11.84IKK501 pKa = 10.82FPLFGEE507 pKa = 4.23EE508 pKa = 4.83VPFLVNGDD516 pKa = 4.53DD517 pKa = 3.92YY518 pKa = 11.94LSFLPVRR525 pKa = 11.84YY526 pKa = 9.6YY527 pKa = 10.25EE528 pKa = 4.02FWKK531 pKa = 10.77EE532 pKa = 4.03CVTSVGLKK540 pKa = 10.06PSLGKK545 pKa = 10.42NYY547 pKa = 9.58LHH549 pKa = 7.57PYY551 pKa = 9.49LGTVNSTLIKK561 pKa = 10.12INRR564 pKa = 11.84PLHH567 pKa = 5.13FDD569 pKa = 3.38EE570 pKa = 5.4RR571 pKa = 11.84FKK573 pKa = 11.71DD574 pKa = 3.7NLPGKK579 pKa = 9.94IFRR582 pKa = 11.84HH583 pKa = 5.54LKK585 pKa = 10.48LSLAYY590 pKa = 9.37PPEE593 pKa = 4.27EE594 pKa = 3.6QDD596 pKa = 4.77VLIKK600 pKa = 10.78DD601 pKa = 3.9DD602 pKa = 3.64TFDD605 pKa = 3.18WRR607 pKa = 11.84KK608 pKa = 10.08ISIKK612 pKa = 10.01TNLEE616 pKa = 3.76RR617 pKa = 11.84LILDD621 pKa = 3.26QKK623 pKa = 11.33EE624 pKa = 4.2DD625 pKa = 3.35IQRR628 pKa = 11.84TLIFYY633 pKa = 10.53ALRR636 pKa = 11.84KK637 pKa = 8.86FKK639 pKa = 10.77KK640 pKa = 9.3FLSVIPKK647 pKa = 8.32EE648 pKa = 4.08IPYY651 pKa = 10.27SVSPSLGGLGIPLVDD666 pKa = 4.01HH667 pKa = 6.47KK668 pKa = 11.08QVSDD672 pKa = 3.46PHH674 pKa = 7.63RR675 pKa = 11.84MRR677 pKa = 11.84VSYY680 pKa = 10.82LITRR684 pKa = 11.84PPKK687 pKa = 10.1RR688 pKa = 11.84GRR690 pKa = 11.84TLIFPGEE697 pKa = 4.2VSQVDD702 pKa = 3.97LLLKK706 pKa = 10.4EE707 pKa = 4.88KK708 pKa = 10.54EE709 pKa = 4.16NQLLDD714 pKa = 3.5QLEE717 pKa = 4.55IPWEE721 pKa = 4.42PGTEE725 pKa = 3.99KK726 pKa = 10.58PSPRR730 pKa = 11.84EE731 pKa = 3.82GEE733 pKa = 4.37SPFSRR738 pKa = 11.84WCFLYY743 pKa = 10.22EE744 pKa = 4.07FEE746 pKa = 5.09KK747 pKa = 10.66IKK749 pKa = 10.7PKK751 pKa = 10.77VSILQTLKK759 pKa = 9.63KK760 pKa = 8.33WSYY763 pKa = 9.61IIKK766 pKa = 10.28RR767 pKa = 11.84EE768 pKa = 4.02LKK770 pKa = 10.09EE771 pKa = 4.17SNRR774 pKa = 11.84LLKK777 pKa = 10.34VLRR780 pKa = 11.84PASSTSCKK788 pKa = 9.89QDD790 pKa = 3.13VKK792 pKa = 10.3PWTRR796 pKa = 11.84TFPLYY801 pKa = 10.62TFDD804 pKa = 5.62LGLGKK809 pKa = 10.25DD810 pKa = 3.91DD811 pKa = 4.08FVSRR815 pKa = 11.84AALFRR820 pKa = 11.84EE821 pKa = 4.09ITPGIDD827 pKa = 3.55LFII830 pKa = 6.14
Molecular weight: 96.6 kDa
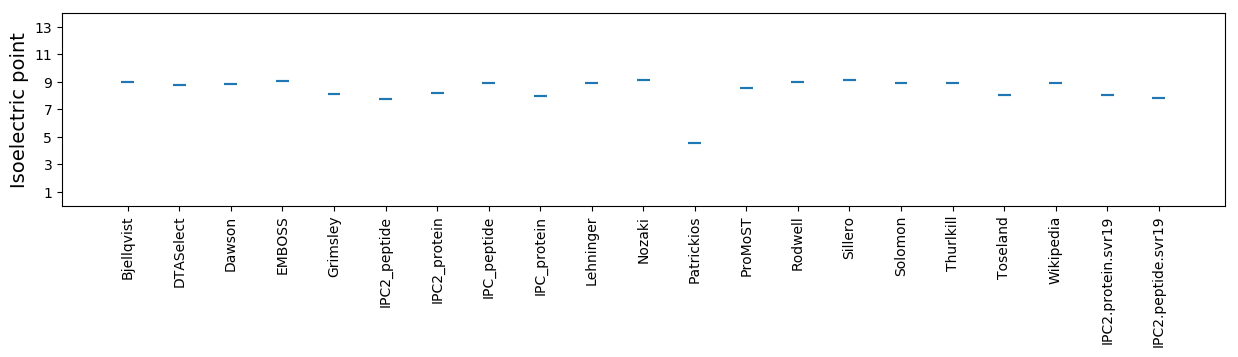
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIU3|A0A1L3KIU3_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 4 OX=1922958 PE=4 SV=1
MM1 pKa = 7.64FIFEE5 pKa = 4.64TFSGQCKK12 pKa = 8.64EE13 pKa = 4.22WQNLCHH19 pKa = 6.45SRR21 pKa = 11.84VKK23 pKa = 10.17EE24 pKa = 3.66ALRR27 pKa = 11.84IRR29 pKa = 11.84KK30 pKa = 8.62LPACWACHH38 pKa = 5.88FPSDD42 pKa = 5.2DD43 pKa = 3.59NPEE46 pKa = 4.02YY47 pKa = 10.56LQVVTSLFNSKK58 pKa = 9.95VEE60 pKa = 4.1EE61 pKa = 4.33KK62 pKa = 10.42DD63 pKa = 3.02WPKK66 pKa = 10.52YY67 pKa = 8.74IQWAIDD73 pKa = 3.47FSTRR77 pKa = 11.84LLEE80 pKa = 4.44SYY82 pKa = 8.96LTYY85 pKa = 10.74FEE87 pKa = 5.78LGLNRR92 pKa = 11.84VLPTDD97 pKa = 3.68RR98 pKa = 11.84KK99 pKa = 9.82ILRR102 pKa = 11.84KK103 pKa = 10.23VIFDD107 pKa = 3.94CLDD110 pKa = 3.11YY111 pKa = 10.99WFLLSLIGKK120 pKa = 8.46LPKK123 pKa = 9.57YY124 pKa = 10.18LKK126 pKa = 10.47YY127 pKa = 7.9WTCKK131 pKa = 9.58FFSDD135 pKa = 4.7ILQQFDD141 pKa = 3.61QPEE144 pKa = 4.22KK145 pKa = 10.89PEE147 pKa = 4.26IPLKK151 pKa = 10.42FKK153 pKa = 10.78PFICGQIAGKK163 pKa = 9.22IHH165 pKa = 7.44LITYY169 pKa = 8.64VRR171 pKa = 11.84TRR173 pKa = 11.84SDD175 pKa = 3.26RR176 pKa = 11.84KK177 pKa = 9.93PSEE180 pKa = 3.77DD181 pKa = 3.29HH182 pKa = 6.64IKK184 pKa = 10.73FFFGLLDD191 pKa = 3.71LKK193 pKa = 10.75KK194 pKa = 10.53ASRR197 pKa = 11.84PIHH200 pKa = 6.16KK201 pKa = 9.92IFVTEE206 pKa = 3.96EE207 pKa = 3.91VEE209 pKa = 4.43KK210 pKa = 10.95SIEE213 pKa = 4.01RR214 pKa = 11.84LTTSPEE220 pKa = 3.86RR221 pKa = 11.84EE222 pKa = 3.9PLRR225 pKa = 11.84NGIIRR230 pKa = 11.84SIKK233 pKa = 9.61RR234 pKa = 11.84VCNSLVFSHH243 pKa = 6.86IRR245 pKa = 11.84GNCSDD250 pKa = 3.86IPINLPSCLPTTGGLEE266 pKa = 3.91NPRR269 pKa = 11.84RR270 pKa = 11.84TGGCYY275 pKa = 10.42GFLQGNLSSLCNNISGSFHH294 pKa = 6.77LCNMEE299 pKa = 3.92KK300 pKa = 10.39DD301 pKa = 4.0GEE303 pKa = 4.2ILPVSVPFNPRR314 pKa = 11.84IEE316 pKa = 4.05LSYY319 pKa = 10.78YY320 pKa = 10.26RR321 pKa = 11.84HH322 pKa = 6.83LDD324 pKa = 3.9DD325 pKa = 5.99LSKK328 pKa = 9.66TYY330 pKa = 10.1PGSIPVEE337 pKa = 4.06MVGLPEE343 pKa = 4.12PFKK346 pKa = 11.32VRR348 pKa = 11.84TISKK352 pKa = 10.7GPIDD356 pKa = 5.32LYY358 pKa = 11.01AQARR362 pKa = 11.84RR363 pKa = 11.84YY364 pKa = 9.71QGLFWHH370 pKa = 7.31LLRR373 pKa = 11.84HH374 pKa = 5.66SPVFSLTWSPLCEE387 pKa = 3.6YY388 pKa = 10.23HH389 pKa = 7.34LRR391 pKa = 11.84YY392 pKa = 9.83LYY394 pKa = 10.77NGNPFSKK401 pKa = 10.49NRR403 pKa = 11.84LEE405 pKa = 4.49SYY407 pKa = 10.43LLSADD412 pKa = 3.72YY413 pKa = 10.42EE414 pKa = 4.21AATDD418 pKa = 4.02SIYY421 pKa = 11.09KK422 pKa = 9.96EE423 pKa = 4.26IIEE426 pKa = 4.28EE427 pKa = 4.12TAEE430 pKa = 4.16HH431 pKa = 6.98LFHH434 pKa = 7.68LLDD437 pKa = 4.31IPWSDD442 pKa = 3.2RR443 pKa = 11.84ALLLKK448 pKa = 10.28ILTRR452 pKa = 11.84CEE454 pKa = 3.48IEE456 pKa = 4.21RR457 pKa = 11.84LDD459 pKa = 4.94GEE461 pKa = 4.38SLKK464 pKa = 10.61QKK466 pKa = 10.17RR467 pKa = 11.84GQLMGSPLSFVLLCIINAGVNLFVIEE493 pKa = 4.14KK494 pKa = 10.5SIGEE498 pKa = 4.15RR499 pKa = 11.84IKK501 pKa = 10.82FPLFGEE507 pKa = 4.23EE508 pKa = 4.83VPFLVNGDD516 pKa = 4.53DD517 pKa = 3.92YY518 pKa = 11.94LSFLPVRR525 pKa = 11.84YY526 pKa = 9.6YY527 pKa = 10.25EE528 pKa = 4.02FWKK531 pKa = 10.77EE532 pKa = 4.03CVTSVGLKK540 pKa = 10.06PSLGKK545 pKa = 10.42NYY547 pKa = 9.58LHH549 pKa = 7.57PYY551 pKa = 9.49LGTVNSTLIKK561 pKa = 10.12INRR564 pKa = 11.84PLHH567 pKa = 5.13FDD569 pKa = 3.38EE570 pKa = 5.4RR571 pKa = 11.84FKK573 pKa = 11.71DD574 pKa = 3.7NLPGKK579 pKa = 9.94IFRR582 pKa = 11.84HH583 pKa = 5.54LKK585 pKa = 10.48LSLAYY590 pKa = 9.37PPEE593 pKa = 4.27EE594 pKa = 3.6QDD596 pKa = 4.77VLIKK600 pKa = 10.78DD601 pKa = 3.9DD602 pKa = 3.64TFDD605 pKa = 3.18WRR607 pKa = 11.84KK608 pKa = 10.08ISIKK612 pKa = 10.01TNLEE616 pKa = 3.76RR617 pKa = 11.84LILDD621 pKa = 3.26QKK623 pKa = 11.33EE624 pKa = 4.2DD625 pKa = 3.35IQRR628 pKa = 11.84TLIFYY633 pKa = 10.53ALRR636 pKa = 11.84KK637 pKa = 8.86FKK639 pKa = 10.77KK640 pKa = 9.3FLSVIPKK647 pKa = 8.32EE648 pKa = 4.08IPYY651 pKa = 10.27SVSPSLGGLGIPLVDD666 pKa = 4.01HH667 pKa = 6.47KK668 pKa = 11.08QVSDD672 pKa = 3.46PHH674 pKa = 7.63RR675 pKa = 11.84MRR677 pKa = 11.84VSYY680 pKa = 10.82LITRR684 pKa = 11.84PPKK687 pKa = 10.1RR688 pKa = 11.84GRR690 pKa = 11.84TLIFPGEE697 pKa = 4.2VSQVDD702 pKa = 3.97LLLKK706 pKa = 10.4EE707 pKa = 4.88KK708 pKa = 10.54EE709 pKa = 4.16NQLLDD714 pKa = 3.5QLEE717 pKa = 4.55IPWEE721 pKa = 4.42PGTEE725 pKa = 3.99KK726 pKa = 10.58PSPRR730 pKa = 11.84EE731 pKa = 3.82GEE733 pKa = 4.37SPFSRR738 pKa = 11.84WCFLYY743 pKa = 10.22EE744 pKa = 4.07FEE746 pKa = 5.09KK747 pKa = 10.66IKK749 pKa = 10.7PKK751 pKa = 10.77VSILQTLKK759 pKa = 9.63KK760 pKa = 8.33WSYY763 pKa = 9.61IIKK766 pKa = 10.28RR767 pKa = 11.84EE768 pKa = 4.02LKK770 pKa = 10.09EE771 pKa = 4.17SNRR774 pKa = 11.84LLKK777 pKa = 10.34VLRR780 pKa = 11.84PASSTSCKK788 pKa = 9.89QDD790 pKa = 3.13VKK792 pKa = 10.3PWTRR796 pKa = 11.84TFPLYY801 pKa = 10.62TFDD804 pKa = 5.62LGLGKK809 pKa = 10.25DD810 pKa = 3.91DD811 pKa = 4.08FVSRR815 pKa = 11.84AALFRR820 pKa = 11.84EE821 pKa = 4.09ITPGIDD827 pKa = 3.55LFII830 pKa = 6.14
MM1 pKa = 7.64FIFEE5 pKa = 4.64TFSGQCKK12 pKa = 8.64EE13 pKa = 4.22WQNLCHH19 pKa = 6.45SRR21 pKa = 11.84VKK23 pKa = 10.17EE24 pKa = 3.66ALRR27 pKa = 11.84IRR29 pKa = 11.84KK30 pKa = 8.62LPACWACHH38 pKa = 5.88FPSDD42 pKa = 5.2DD43 pKa = 3.59NPEE46 pKa = 4.02YY47 pKa = 10.56LQVVTSLFNSKK58 pKa = 9.95VEE60 pKa = 4.1EE61 pKa = 4.33KK62 pKa = 10.42DD63 pKa = 3.02WPKK66 pKa = 10.52YY67 pKa = 8.74IQWAIDD73 pKa = 3.47FSTRR77 pKa = 11.84LLEE80 pKa = 4.44SYY82 pKa = 8.96LTYY85 pKa = 10.74FEE87 pKa = 5.78LGLNRR92 pKa = 11.84VLPTDD97 pKa = 3.68RR98 pKa = 11.84KK99 pKa = 9.82ILRR102 pKa = 11.84KK103 pKa = 10.23VIFDD107 pKa = 3.94CLDD110 pKa = 3.11YY111 pKa = 10.99WFLLSLIGKK120 pKa = 8.46LPKK123 pKa = 9.57YY124 pKa = 10.18LKK126 pKa = 10.47YY127 pKa = 7.9WTCKK131 pKa = 9.58FFSDD135 pKa = 4.7ILQQFDD141 pKa = 3.61QPEE144 pKa = 4.22KK145 pKa = 10.89PEE147 pKa = 4.26IPLKK151 pKa = 10.42FKK153 pKa = 10.78PFICGQIAGKK163 pKa = 9.22IHH165 pKa = 7.44LITYY169 pKa = 8.64VRR171 pKa = 11.84TRR173 pKa = 11.84SDD175 pKa = 3.26RR176 pKa = 11.84KK177 pKa = 9.93PSEE180 pKa = 3.77DD181 pKa = 3.29HH182 pKa = 6.64IKK184 pKa = 10.73FFFGLLDD191 pKa = 3.71LKK193 pKa = 10.75KK194 pKa = 10.53ASRR197 pKa = 11.84PIHH200 pKa = 6.16KK201 pKa = 9.92IFVTEE206 pKa = 3.96EE207 pKa = 3.91VEE209 pKa = 4.43KK210 pKa = 10.95SIEE213 pKa = 4.01RR214 pKa = 11.84LTTSPEE220 pKa = 3.86RR221 pKa = 11.84EE222 pKa = 3.9PLRR225 pKa = 11.84NGIIRR230 pKa = 11.84SIKK233 pKa = 9.61RR234 pKa = 11.84VCNSLVFSHH243 pKa = 6.86IRR245 pKa = 11.84GNCSDD250 pKa = 3.86IPINLPSCLPTTGGLEE266 pKa = 3.91NPRR269 pKa = 11.84RR270 pKa = 11.84TGGCYY275 pKa = 10.42GFLQGNLSSLCNNISGSFHH294 pKa = 6.77LCNMEE299 pKa = 3.92KK300 pKa = 10.39DD301 pKa = 4.0GEE303 pKa = 4.2ILPVSVPFNPRR314 pKa = 11.84IEE316 pKa = 4.05LSYY319 pKa = 10.78YY320 pKa = 10.26RR321 pKa = 11.84HH322 pKa = 6.83LDD324 pKa = 3.9DD325 pKa = 5.99LSKK328 pKa = 9.66TYY330 pKa = 10.1PGSIPVEE337 pKa = 4.06MVGLPEE343 pKa = 4.12PFKK346 pKa = 11.32VRR348 pKa = 11.84TISKK352 pKa = 10.7GPIDD356 pKa = 5.32LYY358 pKa = 11.01AQARR362 pKa = 11.84RR363 pKa = 11.84YY364 pKa = 9.71QGLFWHH370 pKa = 7.31LLRR373 pKa = 11.84HH374 pKa = 5.66SPVFSLTWSPLCEE387 pKa = 3.6YY388 pKa = 10.23HH389 pKa = 7.34LRR391 pKa = 11.84YY392 pKa = 9.83LYY394 pKa = 10.77NGNPFSKK401 pKa = 10.49NRR403 pKa = 11.84LEE405 pKa = 4.49SYY407 pKa = 10.43LLSADD412 pKa = 3.72YY413 pKa = 10.42EE414 pKa = 4.21AATDD418 pKa = 4.02SIYY421 pKa = 11.09KK422 pKa = 9.96EE423 pKa = 4.26IIEE426 pKa = 4.28EE427 pKa = 4.12TAEE430 pKa = 4.16HH431 pKa = 6.98LFHH434 pKa = 7.68LLDD437 pKa = 4.31IPWSDD442 pKa = 3.2RR443 pKa = 11.84ALLLKK448 pKa = 10.28ILTRR452 pKa = 11.84CEE454 pKa = 3.48IEE456 pKa = 4.21RR457 pKa = 11.84LDD459 pKa = 4.94GEE461 pKa = 4.38SLKK464 pKa = 10.61QKK466 pKa = 10.17RR467 pKa = 11.84GQLMGSPLSFVLLCIINAGVNLFVIEE493 pKa = 4.14KK494 pKa = 10.5SIGEE498 pKa = 4.15RR499 pKa = 11.84IKK501 pKa = 10.82FPLFGEE507 pKa = 4.23EE508 pKa = 4.83VPFLVNGDD516 pKa = 4.53DD517 pKa = 3.92YY518 pKa = 11.94LSFLPVRR525 pKa = 11.84YY526 pKa = 9.6YY527 pKa = 10.25EE528 pKa = 4.02FWKK531 pKa = 10.77EE532 pKa = 4.03CVTSVGLKK540 pKa = 10.06PSLGKK545 pKa = 10.42NYY547 pKa = 9.58LHH549 pKa = 7.57PYY551 pKa = 9.49LGTVNSTLIKK561 pKa = 10.12INRR564 pKa = 11.84PLHH567 pKa = 5.13FDD569 pKa = 3.38EE570 pKa = 5.4RR571 pKa = 11.84FKK573 pKa = 11.71DD574 pKa = 3.7NLPGKK579 pKa = 9.94IFRR582 pKa = 11.84HH583 pKa = 5.54LKK585 pKa = 10.48LSLAYY590 pKa = 9.37PPEE593 pKa = 4.27EE594 pKa = 3.6QDD596 pKa = 4.77VLIKK600 pKa = 10.78DD601 pKa = 3.9DD602 pKa = 3.64TFDD605 pKa = 3.18WRR607 pKa = 11.84KK608 pKa = 10.08ISIKK612 pKa = 10.01TNLEE616 pKa = 3.76RR617 pKa = 11.84LILDD621 pKa = 3.26QKK623 pKa = 11.33EE624 pKa = 4.2DD625 pKa = 3.35IQRR628 pKa = 11.84TLIFYY633 pKa = 10.53ALRR636 pKa = 11.84KK637 pKa = 8.86FKK639 pKa = 10.77KK640 pKa = 9.3FLSVIPKK647 pKa = 8.32EE648 pKa = 4.08IPYY651 pKa = 10.27SVSPSLGGLGIPLVDD666 pKa = 4.01HH667 pKa = 6.47KK668 pKa = 11.08QVSDD672 pKa = 3.46PHH674 pKa = 7.63RR675 pKa = 11.84MRR677 pKa = 11.84VSYY680 pKa = 10.82LITRR684 pKa = 11.84PPKK687 pKa = 10.1RR688 pKa = 11.84GRR690 pKa = 11.84TLIFPGEE697 pKa = 4.2VSQVDD702 pKa = 3.97LLLKK706 pKa = 10.4EE707 pKa = 4.88KK708 pKa = 10.54EE709 pKa = 4.16NQLLDD714 pKa = 3.5QLEE717 pKa = 4.55IPWEE721 pKa = 4.42PGTEE725 pKa = 3.99KK726 pKa = 10.58PSPRR730 pKa = 11.84EE731 pKa = 3.82GEE733 pKa = 4.37SPFSRR738 pKa = 11.84WCFLYY743 pKa = 10.22EE744 pKa = 4.07FEE746 pKa = 5.09KK747 pKa = 10.66IKK749 pKa = 10.7PKK751 pKa = 10.77VSILQTLKK759 pKa = 9.63KK760 pKa = 8.33WSYY763 pKa = 9.61IIKK766 pKa = 10.28RR767 pKa = 11.84EE768 pKa = 4.02LKK770 pKa = 10.09EE771 pKa = 4.17SNRR774 pKa = 11.84LLKK777 pKa = 10.34VLRR780 pKa = 11.84PASSTSCKK788 pKa = 9.89QDD790 pKa = 3.13VKK792 pKa = 10.3PWTRR796 pKa = 11.84TFPLYY801 pKa = 10.62TFDD804 pKa = 5.62LGLGKK809 pKa = 10.25DD810 pKa = 3.91DD811 pKa = 4.08FVSRR815 pKa = 11.84AALFRR820 pKa = 11.84EE821 pKa = 4.09ITPGIDD827 pKa = 3.55LFII830 pKa = 6.14
Molecular weight: 96.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
830 |
830 |
830 |
830.0 |
96.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.289 ± 0.0 | 2.289 ± 0.0 |
4.94 ± 0.0 | 6.988 ± 0.0 |
6.024 ± 0.0 | 5.06 ± 0.0 |
2.169 ± 0.0 | 7.47 ± 0.0 |
7.831 ± 0.0 | 13.012 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.602 ± 0.0 | 3.253 ± 0.0 |
6.988 ± 0.0 | 2.651 ± 0.0 |
6.386 ± 0.0 | 7.59 ± 0.0 |
4.217 ± 0.0 | 4.578 ± 0.0 |
1.807 ± 0.0 | 3.855 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
