
Cryptosporangium aurantiacum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Cryptosporangiales; Cryptosporangiaceae; Cryptosporangium
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
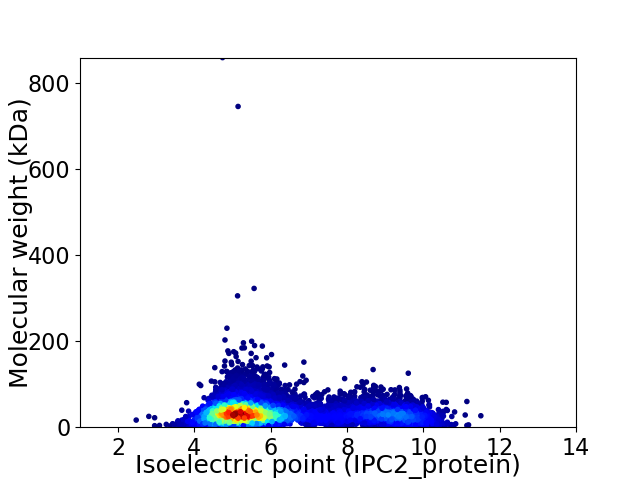
Virtual 2D-PAGE plot for 8789 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M7NP67|A0A1M7NP67_9ACTN Uncharacterized protein OS=Cryptosporangium aurantiacum OX=134849 GN=SAMN05443668_102748 PE=4 SV=1
MM1 pKa = 7.95ADD3 pKa = 5.07FEE5 pKa = 5.3DD6 pKa = 3.88QVDD9 pKa = 3.62WDD11 pKa = 4.91EE12 pKa = 4.06IDD14 pKa = 3.33YY15 pKa = 8.27WTDD18 pKa = 2.71RR19 pKa = 11.84CGACDD24 pKa = 3.74EE25 pKa = 4.69RR26 pKa = 11.84CGGTCEE32 pKa = 5.34DD33 pKa = 4.44YY34 pKa = 11.17PIDD37 pKa = 3.75TTKK40 pKa = 10.74EE41 pKa = 4.09GPRR44 pKa = 3.52
MM1 pKa = 7.95ADD3 pKa = 5.07FEE5 pKa = 5.3DD6 pKa = 3.88QVDD9 pKa = 3.62WDD11 pKa = 4.91EE12 pKa = 4.06IDD14 pKa = 3.33YY15 pKa = 8.27WTDD18 pKa = 2.71RR19 pKa = 11.84CGACDD24 pKa = 3.74EE25 pKa = 4.69RR26 pKa = 11.84CGGTCEE32 pKa = 5.34DD33 pKa = 4.44YY34 pKa = 11.17PIDD37 pKa = 3.75TTKK40 pKa = 10.74EE41 pKa = 4.09GPRR44 pKa = 3.52
Molecular weight: 5.11 kDa
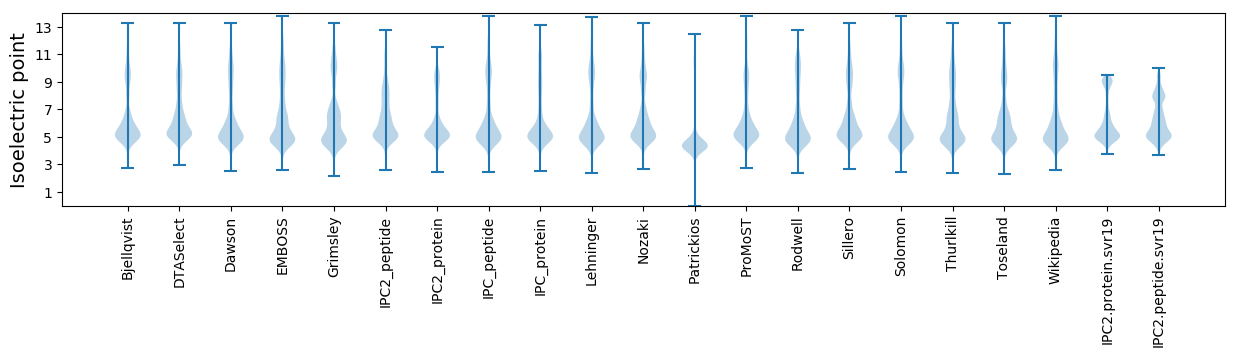
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7RDX3|A0A1M7RDX3_9ACTN VOC domain-containing protein OS=Cryptosporangium aurantiacum OX=134849 GN=SAMN05443668_110200 PE=4 SV=1
MM1 pKa = 8.19DD2 pKa = 5.11IGHH5 pKa = 6.86TPLQCSGQMLGGSNPCKK22 pKa = 10.16APARR26 pKa = 11.84ATVRR30 pKa = 11.84PTVVPVPGRR39 pKa = 11.84TPRR42 pKa = 11.84CPASRR47 pKa = 11.84PRR49 pKa = 11.84SDD51 pKa = 3.38GQSRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.54PGPRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PTVCRR69 pKa = 11.84RR70 pKa = 11.84PSTCSPVPDD79 pKa = 3.88GRR81 pKa = 11.84RR82 pKa = 11.84APPARR87 pKa = 11.84TGSSGSPRR95 pKa = 11.84RR96 pKa = 11.84PRR98 pKa = 11.84CPRR101 pKa = 11.84SRR103 pKa = 11.84ARR105 pKa = 11.84TPRR108 pKa = 11.84PPGRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84ASSRR119 pKa = 11.84RR120 pKa = 11.84CRR122 pKa = 11.84SGRR125 pKa = 11.84GPTAPSRR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 5.37RR135 pKa = 11.84RR136 pKa = 11.84TPGRR140 pKa = 11.84TPRR143 pKa = 11.84PCPAWHH149 pKa = 6.25RR150 pKa = 11.84TSSGSRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84VGRR163 pKa = 11.84VRR165 pKa = 11.84WVPVTLTRR173 pKa = 11.84RR174 pKa = 11.84TSSGSASSAIRR185 pKa = 11.84RR186 pKa = 11.84PRR188 pKa = 11.84TGRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84QRR195 pKa = 11.84PVPSPSNSRR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84SRR208 pKa = 11.84SPVANHH214 pKa = 6.93RR215 pKa = 11.84GRR217 pKa = 11.84ARR219 pKa = 11.84SSRR222 pKa = 11.84PAANRR227 pKa = 11.84SGQAQSSRR235 pKa = 11.84PAANRR240 pKa = 11.84SGRR243 pKa = 11.84ARR245 pKa = 11.84SSRR248 pKa = 11.84PVASRR253 pKa = 11.84RR254 pKa = 11.84GRR256 pKa = 11.84ARR258 pKa = 11.84SSTRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84ARR266 pKa = 11.84SSSGRR271 pKa = 11.84GSTSRR276 pKa = 11.84RR277 pKa = 11.84GSTSRR282 pKa = 11.84RR283 pKa = 11.84GSTSRR288 pKa = 11.84RR289 pKa = 11.84GSTSGPRR296 pKa = 11.84SRR298 pKa = 11.84RR299 pKa = 11.84ASSGRR304 pKa = 11.84RR305 pKa = 11.84SSHH308 pKa = 6.07RR309 pKa = 11.84ASSSGRR315 pKa = 11.84RR316 pKa = 11.84SNRR319 pKa = 11.84TNNSNGPPSSRR330 pKa = 11.84RR331 pKa = 11.84ASSSGRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84HH340 pKa = 5.39RR341 pKa = 11.84TTSSSGRR348 pKa = 11.84RR349 pKa = 11.84SSNRR353 pKa = 11.84RR354 pKa = 11.84SGRR357 pKa = 11.84SSSPAGRR364 pKa = 11.84RR365 pKa = 11.84RR366 pKa = 11.84NRR368 pKa = 11.84SGPRR372 pKa = 11.84NSSGPARR379 pKa = 11.84RR380 pKa = 11.84TSGLPSRR387 pKa = 11.84SGPRR391 pKa = 11.84SSSGRR396 pKa = 11.84GRR398 pKa = 11.84RR399 pKa = 11.84TSGPRR404 pKa = 11.84RR405 pKa = 11.84SSSNGLRR412 pKa = 11.84SSHH415 pKa = 6.46SGPPSSSPRR424 pKa = 11.84RR425 pKa = 11.84RR426 pKa = 11.84PTSRR430 pKa = 11.84PSPSSPTSPSNGRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84SSSGPHH451 pKa = 5.0PRR453 pKa = 11.84IRR455 pKa = 11.84TTRR458 pKa = 11.84SPGSRR463 pKa = 11.84PPSRR467 pKa = 11.84AGSRR471 pKa = 11.84SPATPRR477 pKa = 11.84PPRR480 pKa = 11.84RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84DD484 pKa = 3.36SRR486 pKa = 11.84SPASPCTTTRR496 pKa = 11.84YY497 pKa = 9.45PGSPSPGSPSPGSPSPGSPSPGSPSPGSRR526 pKa = 11.84CTTTRR531 pKa = 11.84YY532 pKa = 9.41PGSRR536 pKa = 11.84PTGSPSRR543 pKa = 11.84GSPSRR548 pKa = 11.84GSTLSPTRR556 pKa = 3.68
MM1 pKa = 8.19DD2 pKa = 5.11IGHH5 pKa = 6.86TPLQCSGQMLGGSNPCKK22 pKa = 10.16APARR26 pKa = 11.84ATVRR30 pKa = 11.84PTVVPVPGRR39 pKa = 11.84TPRR42 pKa = 11.84CPASRR47 pKa = 11.84PRR49 pKa = 11.84SDD51 pKa = 3.38GQSRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.54PGPRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PTVCRR69 pKa = 11.84RR70 pKa = 11.84PSTCSPVPDD79 pKa = 3.88GRR81 pKa = 11.84RR82 pKa = 11.84APPARR87 pKa = 11.84TGSSGSPRR95 pKa = 11.84RR96 pKa = 11.84PRR98 pKa = 11.84CPRR101 pKa = 11.84SRR103 pKa = 11.84ARR105 pKa = 11.84TPRR108 pKa = 11.84PPGRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84ASSRR119 pKa = 11.84RR120 pKa = 11.84CRR122 pKa = 11.84SGRR125 pKa = 11.84GPTAPSRR132 pKa = 11.84RR133 pKa = 11.84HH134 pKa = 5.37RR135 pKa = 11.84RR136 pKa = 11.84TPGRR140 pKa = 11.84TPRR143 pKa = 11.84PCPAWHH149 pKa = 6.25RR150 pKa = 11.84TSSGSRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84VGRR163 pKa = 11.84VRR165 pKa = 11.84WVPVTLTRR173 pKa = 11.84RR174 pKa = 11.84TSSGSASSAIRR185 pKa = 11.84RR186 pKa = 11.84PRR188 pKa = 11.84TGRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84QRR195 pKa = 11.84PVPSPSNSRR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84SRR208 pKa = 11.84SPVANHH214 pKa = 6.93RR215 pKa = 11.84GRR217 pKa = 11.84ARR219 pKa = 11.84SSRR222 pKa = 11.84PAANRR227 pKa = 11.84SGQAQSSRR235 pKa = 11.84PAANRR240 pKa = 11.84SGRR243 pKa = 11.84ARR245 pKa = 11.84SSRR248 pKa = 11.84PVASRR253 pKa = 11.84RR254 pKa = 11.84GRR256 pKa = 11.84ARR258 pKa = 11.84SSTRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84ARR266 pKa = 11.84SSSGRR271 pKa = 11.84GSTSRR276 pKa = 11.84RR277 pKa = 11.84GSTSRR282 pKa = 11.84RR283 pKa = 11.84GSTSRR288 pKa = 11.84RR289 pKa = 11.84GSTSGPRR296 pKa = 11.84SRR298 pKa = 11.84RR299 pKa = 11.84ASSGRR304 pKa = 11.84RR305 pKa = 11.84SSHH308 pKa = 6.07RR309 pKa = 11.84ASSSGRR315 pKa = 11.84RR316 pKa = 11.84SNRR319 pKa = 11.84TNNSNGPPSSRR330 pKa = 11.84RR331 pKa = 11.84ASSSGRR337 pKa = 11.84RR338 pKa = 11.84RR339 pKa = 11.84HH340 pKa = 5.39RR341 pKa = 11.84TTSSSGRR348 pKa = 11.84RR349 pKa = 11.84SSNRR353 pKa = 11.84RR354 pKa = 11.84SGRR357 pKa = 11.84SSSPAGRR364 pKa = 11.84RR365 pKa = 11.84RR366 pKa = 11.84NRR368 pKa = 11.84SGPRR372 pKa = 11.84NSSGPARR379 pKa = 11.84RR380 pKa = 11.84TSGLPSRR387 pKa = 11.84SGPRR391 pKa = 11.84SSSGRR396 pKa = 11.84GRR398 pKa = 11.84RR399 pKa = 11.84TSGPRR404 pKa = 11.84RR405 pKa = 11.84SSSNGLRR412 pKa = 11.84SSHH415 pKa = 6.46SGPPSSSPRR424 pKa = 11.84RR425 pKa = 11.84RR426 pKa = 11.84PTSRR430 pKa = 11.84PSPSSPTSPSNGRR443 pKa = 11.84RR444 pKa = 11.84RR445 pKa = 11.84SSSGPHH451 pKa = 5.0PRR453 pKa = 11.84IRR455 pKa = 11.84TTRR458 pKa = 11.84SPGSRR463 pKa = 11.84PPSRR467 pKa = 11.84AGSRR471 pKa = 11.84SPATPRR477 pKa = 11.84PPRR480 pKa = 11.84RR481 pKa = 11.84RR482 pKa = 11.84RR483 pKa = 11.84DD484 pKa = 3.36SRR486 pKa = 11.84SPASPCTTTRR496 pKa = 11.84YY497 pKa = 9.45PGSPSPGSPSPGSPSPGSPSPGSPSPGSRR526 pKa = 11.84CTTTRR531 pKa = 11.84YY532 pKa = 9.41PGSRR536 pKa = 11.84PTGSPSRR543 pKa = 11.84GSPSRR548 pKa = 11.84GSTLSPTRR556 pKa = 3.68
Molecular weight: 59.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2911538 |
24 |
8163 |
331.3 |
35.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.174 ± 0.044 | 0.695 ± 0.007 |
6.157 ± 0.02 | 5.265 ± 0.024 |
2.749 ± 0.016 | 9.185 ± 0.027 |
2.032 ± 0.012 | 3.41 ± 0.018 |
1.652 ± 0.018 | 10.575 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.513 ± 0.009 | 1.728 ± 0.014 |
6.241 ± 0.024 | 2.585 ± 0.015 |
8.131 ± 0.03 | 5.076 ± 0.017 |
6.151 ± 0.022 | 9.129 ± 0.028 |
1.539 ± 0.01 | 2.013 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
