
Kriegella aquimaris
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Kriegella
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
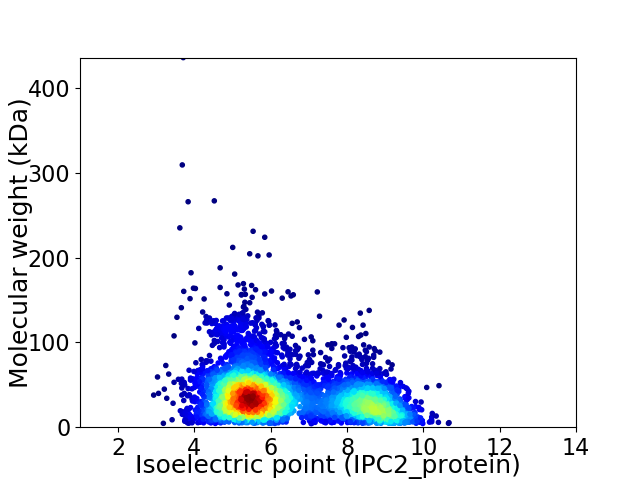
Virtual 2D-PAGE plot for 4956 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9IA92|A0A1G9IA92_9FLAO PhnB protein OS=Kriegella aquimaris OX=192904 GN=SAMN04488514_10157 PE=4 SV=1
MM1 pKa = 7.68FEE3 pKa = 4.7TILFNPTMTYY13 pKa = 9.4TRR15 pKa = 11.84INRR18 pKa = 11.84FFPRR22 pKa = 11.84LNLDD26 pKa = 3.09FTLALRR32 pKa = 11.84FSVKK36 pKa = 10.56ALLCFGMMSLSTNWVAGQEE55 pKa = 4.26TYY57 pKa = 10.88RR58 pKa = 11.84DD59 pKa = 3.57NFSSVNYY66 pKa = 9.96GNNNGSLNFSTDD78 pKa = 2.99WIEE81 pKa = 4.53TGDD84 pKa = 3.71TDD86 pKa = 5.07GGPSSQYY93 pKa = 10.59IRR95 pKa = 11.84ISSNRR100 pKa = 11.84LEE102 pKa = 4.72LYY104 pKa = 10.03YY105 pKa = 10.58LYY107 pKa = 10.75SEE109 pKa = 4.97TIRR112 pKa = 11.84RR113 pKa = 11.84SADD116 pKa = 3.05LSGASTATLSFNWQAISLGGNRR138 pKa = 11.84SLAVQVSNNGGASYY152 pKa = 8.96ATVGTITGNNSSGTFSEE169 pKa = 5.63DD170 pKa = 2.33ISAYY174 pKa = 9.7ISATTTIRR182 pKa = 11.84FRR184 pKa = 11.84KK185 pKa = 9.98SNSNWRR191 pKa = 11.84NDD193 pKa = 3.27DD194 pKa = 3.43YY195 pKa = 12.18AFIDD199 pKa = 3.64NLQISATFPAPIPIIEE215 pKa = 4.2VDD217 pKa = 3.96DD218 pKa = 3.87VTVDD222 pKa = 3.43EE223 pKa = 5.53DD224 pKa = 3.5GGNAIFTVSHH234 pKa = 6.8TGTNAAGAFTVNYY247 pKa = 7.15QTINGSANAGSDD259 pKa = 3.55YY260 pKa = 8.09TTTTGVLNFNGNSGDD275 pKa = 3.71TEE277 pKa = 4.47TFSVPILDD285 pKa = 4.87DD286 pKa = 3.46ATIEE290 pKa = 4.06NAEE293 pKa = 4.31DD294 pKa = 3.54FTIQFLAVSDD304 pKa = 4.01PTVDD308 pKa = 3.17ITDD311 pKa = 3.76TAIGTIVDD319 pKa = 4.01DD320 pKa = 5.12DD321 pKa = 4.49ALIMIDD327 pKa = 3.72GGSATSCSDD336 pKa = 2.69IFFDD340 pKa = 5.51PGGLSNYY347 pKa = 10.05SNNEE351 pKa = 3.66DD352 pKa = 3.07VTYY355 pKa = 8.99TICPDD360 pKa = 3.37TADD363 pKa = 3.45TYY365 pKa = 11.57ININFSTFEE374 pKa = 4.18VVSGDD379 pKa = 3.2VLYY382 pKa = 10.11IYY384 pKa = 10.47EE385 pKa = 4.37GTTTGGTLIGQYY397 pKa = 10.86DD398 pKa = 3.95SANIPTAINSSHH410 pKa = 7.15ASGCLTFRR418 pKa = 11.84FTSNGDD424 pKa = 3.18TTGAGWEE431 pKa = 3.98AAVNCFPEE439 pKa = 4.96GPIIIIDD446 pKa = 4.64DD447 pKa = 3.4ISFDD451 pKa = 3.89EE452 pKa = 5.39DD453 pKa = 2.77IGNAVFTVRR462 pKa = 11.84STRR465 pKa = 11.84AAHH468 pKa = 5.3GTNVFLLGFVNQSFTVDD485 pKa = 4.1FQTVDD490 pKa = 3.19GSALAGSDD498 pKa = 3.45YY499 pKa = 7.98TTTSGTLTFTGALNNVQTISVPIANDD525 pKa = 3.75GVPEE529 pKa = 3.98FAEE532 pKa = 4.3DD533 pKa = 3.56FTIEE537 pKa = 3.96FTGANAQYY545 pKa = 9.93ATVNYY550 pKa = 8.69TDD552 pKa = 3.79TGIGTINSQILANDD566 pKa = 4.05PLTLFQEE573 pKa = 4.19FDD575 pKa = 3.71GYY577 pKa = 11.39FDD579 pKa = 3.64YY580 pKa = 8.05TTTGGSLRR588 pKa = 11.84TSGNGGDD595 pKa = 3.67PCAITTSSTNRR606 pKa = 11.84LVSPIPNTAAIEE618 pKa = 3.89RR619 pKa = 11.84AYY621 pKa = 10.76LYY623 pKa = 7.8WAHH626 pKa = 6.65SNTVRR631 pKa = 11.84DD632 pKa = 4.03DD633 pKa = 3.52NVTFEE638 pKa = 4.34GQNVSANFLYY648 pKa = 8.95QTSLTNRR655 pKa = 11.84NFYY658 pKa = 10.97GYY660 pKa = 10.55VSDD663 pKa = 3.78VTSIVAAIPNISTNDD678 pKa = 3.16FDD680 pKa = 6.83FSDD683 pKa = 4.77LDD685 pKa = 3.38IDD687 pKa = 3.99NTGSYY692 pKa = 10.49CSTATVLGGWSLIVFYY708 pKa = 10.32EE709 pKa = 4.03DD710 pKa = 3.34RR711 pKa = 11.84SLPAVNINLYY721 pKa = 10.54QGFDD725 pKa = 3.45GLSNDD730 pKa = 3.88GNSFTLDD737 pKa = 2.87SFYY740 pKa = 11.09AIAGAGAKK748 pKa = 8.67STFLSWEE755 pKa = 4.11GDD757 pKa = 3.33PDD759 pKa = 3.97LASGGANPEE768 pKa = 3.87RR769 pKa = 11.84LTITNQANTTSNLTGDD785 pKa = 4.32GGQTGTNAYY794 pKa = 10.72NSTIYY799 pKa = 10.85DD800 pKa = 3.56NTVGPVYY807 pKa = 9.69NTSNIYY813 pKa = 10.55GVDD816 pKa = 3.72LDD818 pKa = 4.53TYY820 pKa = 10.17DD821 pKa = 3.03ISSFISPGDD830 pKa = 3.67SQVTANVNVGQDD842 pKa = 3.69FVISAAVVLKK852 pKa = 10.96VPSNLIAGRR861 pKa = 11.84VFEE864 pKa = 4.59DD865 pKa = 3.22VNYY868 pKa = 10.2PGGAGRR874 pKa = 11.84DD875 pKa = 3.57QATSGGIGISGAIVEE890 pKa = 5.04LFDD893 pKa = 4.37SSGNFLQRR901 pKa = 11.84KK902 pKa = 4.49TTNITGNYY910 pKa = 8.89SFGGMADD917 pKa = 2.99GDD919 pKa = 4.26FNVKK923 pKa = 9.05VVNSTVRR930 pKa = 11.84SNRR933 pKa = 11.84NNGLNCSACYY943 pKa = 9.51PIQTFRR949 pKa = 11.84TYY951 pKa = 11.52SDD953 pKa = 2.97VDD955 pKa = 4.01SNLFEE960 pKa = 4.71VINEE964 pKa = 3.96IGGANPNATQDD975 pKa = 3.44VALGIINGAQSVSTVSVASNGVVDD999 pKa = 4.31MDD1001 pKa = 4.09FGFNFSTVVNTNDD1014 pKa = 3.34AGQGSLAQFVVNSNNLGEE1032 pKa = 4.58TILDD1036 pKa = 3.56IEE1038 pKa = 4.84PNSIFDD1044 pKa = 3.81PAAEE1048 pKa = 3.98EE1049 pKa = 4.1DD1050 pKa = 3.78VSIFMIPPTGDD1061 pKa = 3.34PLGRR1065 pKa = 11.84TADD1068 pKa = 3.74TNFSGGIFDD1077 pKa = 5.05ISISNINTITPITGTNTSIDD1097 pKa = 3.27GRR1099 pKa = 11.84TQTAYY1104 pKa = 10.74SGNTNSGTVGSGGTVVGVSANSLPNFDD1131 pKa = 3.95RR1132 pKa = 11.84PVIQVHH1138 pKa = 6.39RR1139 pKa = 11.84NDD1141 pKa = 3.62GDD1143 pKa = 3.79VLEE1146 pKa = 4.6LQGNNTLIRR1155 pKa = 11.84GLSIYY1160 pKa = 10.64ADD1162 pKa = 3.77SNAGILVSGGSSNIIGNLLGVNASGVAAGNIDD1194 pKa = 3.83YY1195 pKa = 10.67GVEE1198 pKa = 3.94IVGGTTTIDD1207 pKa = 3.03GNYY1210 pKa = 9.65ISGNTDD1216 pKa = 2.89EE1217 pKa = 5.79GVLIDD1222 pKa = 4.6GGTSVLVQNNHH1233 pKa = 4.13ITEE1236 pKa = 4.49NGNTACSDD1244 pKa = 3.79NIRR1247 pKa = 11.84IEE1249 pKa = 4.35GGSGIVIQGNLIEE1262 pKa = 4.53NAAALGIDD1270 pKa = 3.86EE1271 pKa = 5.15NGSIGDD1277 pKa = 3.82IQISDD1282 pKa = 3.57NTIVGSGQNGGDD1294 pKa = 3.32CSGLVKK1300 pKa = 10.57SAGIRR1305 pKa = 11.84LEE1307 pKa = 4.3GNNSVVNDD1315 pKa = 4.15NIITSNGGAGIVLVGGNTSGNLISQNSIYY1344 pKa = 11.21ANGTITASLGIDD1356 pKa = 3.01IDD1358 pKa = 3.8QTNQMGDD1365 pKa = 3.17GVTLNEE1371 pKa = 4.88TGDD1374 pKa = 3.83ADD1376 pKa = 3.7NGPNGVINFPIISGVYY1392 pKa = 9.43ISSTNLTIEE1401 pKa = 4.06GWSRR1405 pKa = 11.84PGATIEE1411 pKa = 5.33LFLTDD1416 pKa = 3.68ISEE1419 pKa = 4.54GSAVAGDD1426 pKa = 3.78NEE1428 pKa = 4.57LGKK1431 pKa = 7.78TTDD1434 pKa = 3.47YY1435 pKa = 11.9GEE1437 pKa = 4.48GQAYY1441 pKa = 9.39LASFVEE1447 pKa = 5.02GSGSDD1452 pKa = 3.78LLSDD1456 pKa = 3.64ATAYY1460 pKa = 10.86LDD1462 pKa = 3.49EE1463 pKa = 5.99DD1464 pKa = 4.65GNVDD1468 pKa = 3.42TTNKK1472 pKa = 9.86FKK1474 pKa = 10.3FTIPLPPGVIKK1485 pKa = 10.96GNLVTSTATISNSTSEE1501 pKa = 4.49FSPMSIIKK1509 pKa = 9.29TYY1511 pKa = 8.8TVITNRR1517 pKa = 11.84RR1518 pKa = 11.84ITYY1521 pKa = 10.01RR1522 pKa = 11.84IKK1524 pKa = 10.96KK1525 pKa = 8.93NN1526 pKa = 3.17
MM1 pKa = 7.68FEE3 pKa = 4.7TILFNPTMTYY13 pKa = 9.4TRR15 pKa = 11.84INRR18 pKa = 11.84FFPRR22 pKa = 11.84LNLDD26 pKa = 3.09FTLALRR32 pKa = 11.84FSVKK36 pKa = 10.56ALLCFGMMSLSTNWVAGQEE55 pKa = 4.26TYY57 pKa = 10.88RR58 pKa = 11.84DD59 pKa = 3.57NFSSVNYY66 pKa = 9.96GNNNGSLNFSTDD78 pKa = 2.99WIEE81 pKa = 4.53TGDD84 pKa = 3.71TDD86 pKa = 5.07GGPSSQYY93 pKa = 10.59IRR95 pKa = 11.84ISSNRR100 pKa = 11.84LEE102 pKa = 4.72LYY104 pKa = 10.03YY105 pKa = 10.58LYY107 pKa = 10.75SEE109 pKa = 4.97TIRR112 pKa = 11.84RR113 pKa = 11.84SADD116 pKa = 3.05LSGASTATLSFNWQAISLGGNRR138 pKa = 11.84SLAVQVSNNGGASYY152 pKa = 8.96ATVGTITGNNSSGTFSEE169 pKa = 5.63DD170 pKa = 2.33ISAYY174 pKa = 9.7ISATTTIRR182 pKa = 11.84FRR184 pKa = 11.84KK185 pKa = 9.98SNSNWRR191 pKa = 11.84NDD193 pKa = 3.27DD194 pKa = 3.43YY195 pKa = 12.18AFIDD199 pKa = 3.64NLQISATFPAPIPIIEE215 pKa = 4.2VDD217 pKa = 3.96DD218 pKa = 3.87VTVDD222 pKa = 3.43EE223 pKa = 5.53DD224 pKa = 3.5GGNAIFTVSHH234 pKa = 6.8TGTNAAGAFTVNYY247 pKa = 7.15QTINGSANAGSDD259 pKa = 3.55YY260 pKa = 8.09TTTTGVLNFNGNSGDD275 pKa = 3.71TEE277 pKa = 4.47TFSVPILDD285 pKa = 4.87DD286 pKa = 3.46ATIEE290 pKa = 4.06NAEE293 pKa = 4.31DD294 pKa = 3.54FTIQFLAVSDD304 pKa = 4.01PTVDD308 pKa = 3.17ITDD311 pKa = 3.76TAIGTIVDD319 pKa = 4.01DD320 pKa = 5.12DD321 pKa = 4.49ALIMIDD327 pKa = 3.72GGSATSCSDD336 pKa = 2.69IFFDD340 pKa = 5.51PGGLSNYY347 pKa = 10.05SNNEE351 pKa = 3.66DD352 pKa = 3.07VTYY355 pKa = 8.99TICPDD360 pKa = 3.37TADD363 pKa = 3.45TYY365 pKa = 11.57ININFSTFEE374 pKa = 4.18VVSGDD379 pKa = 3.2VLYY382 pKa = 10.11IYY384 pKa = 10.47EE385 pKa = 4.37GTTTGGTLIGQYY397 pKa = 10.86DD398 pKa = 3.95SANIPTAINSSHH410 pKa = 7.15ASGCLTFRR418 pKa = 11.84FTSNGDD424 pKa = 3.18TTGAGWEE431 pKa = 3.98AAVNCFPEE439 pKa = 4.96GPIIIIDD446 pKa = 4.64DD447 pKa = 3.4ISFDD451 pKa = 3.89EE452 pKa = 5.39DD453 pKa = 2.77IGNAVFTVRR462 pKa = 11.84STRR465 pKa = 11.84AAHH468 pKa = 5.3GTNVFLLGFVNQSFTVDD485 pKa = 4.1FQTVDD490 pKa = 3.19GSALAGSDD498 pKa = 3.45YY499 pKa = 7.98TTTSGTLTFTGALNNVQTISVPIANDD525 pKa = 3.75GVPEE529 pKa = 3.98FAEE532 pKa = 4.3DD533 pKa = 3.56FTIEE537 pKa = 3.96FTGANAQYY545 pKa = 9.93ATVNYY550 pKa = 8.69TDD552 pKa = 3.79TGIGTINSQILANDD566 pKa = 4.05PLTLFQEE573 pKa = 4.19FDD575 pKa = 3.71GYY577 pKa = 11.39FDD579 pKa = 3.64YY580 pKa = 8.05TTTGGSLRR588 pKa = 11.84TSGNGGDD595 pKa = 3.67PCAITTSSTNRR606 pKa = 11.84LVSPIPNTAAIEE618 pKa = 3.89RR619 pKa = 11.84AYY621 pKa = 10.76LYY623 pKa = 7.8WAHH626 pKa = 6.65SNTVRR631 pKa = 11.84DD632 pKa = 4.03DD633 pKa = 3.52NVTFEE638 pKa = 4.34GQNVSANFLYY648 pKa = 8.95QTSLTNRR655 pKa = 11.84NFYY658 pKa = 10.97GYY660 pKa = 10.55VSDD663 pKa = 3.78VTSIVAAIPNISTNDD678 pKa = 3.16FDD680 pKa = 6.83FSDD683 pKa = 4.77LDD685 pKa = 3.38IDD687 pKa = 3.99NTGSYY692 pKa = 10.49CSTATVLGGWSLIVFYY708 pKa = 10.32EE709 pKa = 4.03DD710 pKa = 3.34RR711 pKa = 11.84SLPAVNINLYY721 pKa = 10.54QGFDD725 pKa = 3.45GLSNDD730 pKa = 3.88GNSFTLDD737 pKa = 2.87SFYY740 pKa = 11.09AIAGAGAKK748 pKa = 8.67STFLSWEE755 pKa = 4.11GDD757 pKa = 3.33PDD759 pKa = 3.97LASGGANPEE768 pKa = 3.87RR769 pKa = 11.84LTITNQANTTSNLTGDD785 pKa = 4.32GGQTGTNAYY794 pKa = 10.72NSTIYY799 pKa = 10.85DD800 pKa = 3.56NTVGPVYY807 pKa = 9.69NTSNIYY813 pKa = 10.55GVDD816 pKa = 3.72LDD818 pKa = 4.53TYY820 pKa = 10.17DD821 pKa = 3.03ISSFISPGDD830 pKa = 3.67SQVTANVNVGQDD842 pKa = 3.69FVISAAVVLKK852 pKa = 10.96VPSNLIAGRR861 pKa = 11.84VFEE864 pKa = 4.59DD865 pKa = 3.22VNYY868 pKa = 10.2PGGAGRR874 pKa = 11.84DD875 pKa = 3.57QATSGGIGISGAIVEE890 pKa = 5.04LFDD893 pKa = 4.37SSGNFLQRR901 pKa = 11.84KK902 pKa = 4.49TTNITGNYY910 pKa = 8.89SFGGMADD917 pKa = 2.99GDD919 pKa = 4.26FNVKK923 pKa = 9.05VVNSTVRR930 pKa = 11.84SNRR933 pKa = 11.84NNGLNCSACYY943 pKa = 9.51PIQTFRR949 pKa = 11.84TYY951 pKa = 11.52SDD953 pKa = 2.97VDD955 pKa = 4.01SNLFEE960 pKa = 4.71VINEE964 pKa = 3.96IGGANPNATQDD975 pKa = 3.44VALGIINGAQSVSTVSVASNGVVDD999 pKa = 4.31MDD1001 pKa = 4.09FGFNFSTVVNTNDD1014 pKa = 3.34AGQGSLAQFVVNSNNLGEE1032 pKa = 4.58TILDD1036 pKa = 3.56IEE1038 pKa = 4.84PNSIFDD1044 pKa = 3.81PAAEE1048 pKa = 3.98EE1049 pKa = 4.1DD1050 pKa = 3.78VSIFMIPPTGDD1061 pKa = 3.34PLGRR1065 pKa = 11.84TADD1068 pKa = 3.74TNFSGGIFDD1077 pKa = 5.05ISISNINTITPITGTNTSIDD1097 pKa = 3.27GRR1099 pKa = 11.84TQTAYY1104 pKa = 10.74SGNTNSGTVGSGGTVVGVSANSLPNFDD1131 pKa = 3.95RR1132 pKa = 11.84PVIQVHH1138 pKa = 6.39RR1139 pKa = 11.84NDD1141 pKa = 3.62GDD1143 pKa = 3.79VLEE1146 pKa = 4.6LQGNNTLIRR1155 pKa = 11.84GLSIYY1160 pKa = 10.64ADD1162 pKa = 3.77SNAGILVSGGSSNIIGNLLGVNASGVAAGNIDD1194 pKa = 3.83YY1195 pKa = 10.67GVEE1198 pKa = 3.94IVGGTTTIDD1207 pKa = 3.03GNYY1210 pKa = 9.65ISGNTDD1216 pKa = 2.89EE1217 pKa = 5.79GVLIDD1222 pKa = 4.6GGTSVLVQNNHH1233 pKa = 4.13ITEE1236 pKa = 4.49NGNTACSDD1244 pKa = 3.79NIRR1247 pKa = 11.84IEE1249 pKa = 4.35GGSGIVIQGNLIEE1262 pKa = 4.53NAAALGIDD1270 pKa = 3.86EE1271 pKa = 5.15NGSIGDD1277 pKa = 3.82IQISDD1282 pKa = 3.57NTIVGSGQNGGDD1294 pKa = 3.32CSGLVKK1300 pKa = 10.57SAGIRR1305 pKa = 11.84LEE1307 pKa = 4.3GNNSVVNDD1315 pKa = 4.15NIITSNGGAGIVLVGGNTSGNLISQNSIYY1344 pKa = 11.21ANGTITASLGIDD1356 pKa = 3.01IDD1358 pKa = 3.8QTNQMGDD1365 pKa = 3.17GVTLNEE1371 pKa = 4.88TGDD1374 pKa = 3.83ADD1376 pKa = 3.7NGPNGVINFPIISGVYY1392 pKa = 9.43ISSTNLTIEE1401 pKa = 4.06GWSRR1405 pKa = 11.84PGATIEE1411 pKa = 5.33LFLTDD1416 pKa = 3.68ISEE1419 pKa = 4.54GSAVAGDD1426 pKa = 3.78NEE1428 pKa = 4.57LGKK1431 pKa = 7.78TTDD1434 pKa = 3.47YY1435 pKa = 11.9GEE1437 pKa = 4.48GQAYY1441 pKa = 9.39LASFVEE1447 pKa = 5.02GSGSDD1452 pKa = 3.78LLSDD1456 pKa = 3.64ATAYY1460 pKa = 10.86LDD1462 pKa = 3.49EE1463 pKa = 5.99DD1464 pKa = 4.65GNVDD1468 pKa = 3.42TTNKK1472 pKa = 9.86FKK1474 pKa = 10.3FTIPLPPGVIKK1485 pKa = 10.96GNLVTSTATISNSTSEE1501 pKa = 4.49FSPMSIIKK1509 pKa = 9.29TYY1511 pKa = 8.8TVITNRR1517 pKa = 11.84RR1518 pKa = 11.84ITYY1521 pKa = 10.01RR1522 pKa = 11.84IKK1524 pKa = 10.96KK1525 pKa = 8.93NN1526 pKa = 3.17
Molecular weight: 160.27 kDa
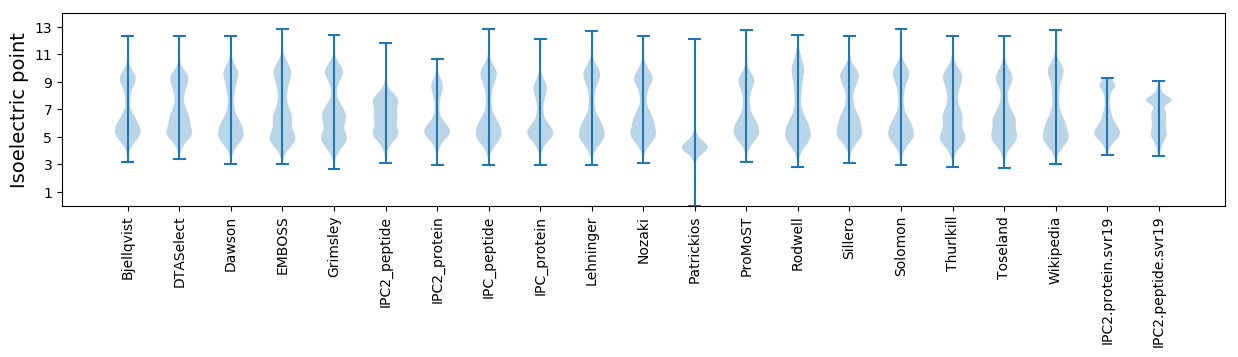
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9TGQ3|A0A1G9TGQ3_9FLAO Quinol:cytochrome c oxidoreductase monoheme cytochrome subunit OS=Kriegella aquimaris OX=192904 GN=SAMN04488514_10959 PE=4 SV=1
MM1 pKa = 7.55DD2 pKa = 3.64VSYY5 pKa = 11.25RR6 pKa = 11.84SMDD9 pKa = 3.04ILFLSRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.33FKK19 pKa = 11.25KK20 pKa = 8.21NTCRR24 pKa = 11.84NAIFKK29 pKa = 10.55KK30 pKa = 10.61GILLEE35 pKa = 4.14EE36 pKa = 4.34TQKK39 pKa = 10.77KK40 pKa = 6.88QHH42 pKa = 6.59RR43 pKa = 11.84LNQTMLFLTNLTNFFVNTPCRR64 pKa = 11.84WRR66 pKa = 11.84ALMLL70 pKa = 4.36
MM1 pKa = 7.55DD2 pKa = 3.64VSYY5 pKa = 11.25RR6 pKa = 11.84SMDD9 pKa = 3.04ILFLSRR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.33FKK19 pKa = 11.25KK20 pKa = 8.21NTCRR24 pKa = 11.84NAIFKK29 pKa = 10.55KK30 pKa = 10.61GILLEE35 pKa = 4.14EE36 pKa = 4.34TQKK39 pKa = 10.77KK40 pKa = 6.88QHH42 pKa = 6.59RR43 pKa = 11.84LNQTMLFLTNLTNFFVNTPCRR64 pKa = 11.84WRR66 pKa = 11.84ALMLL70 pKa = 4.36
Molecular weight: 8.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1798520 |
39 |
4105 |
362.9 |
40.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.598 ± 0.029 | 0.73 ± 0.011 |
5.839 ± 0.025 | 6.534 ± 0.03 |
5.049 ± 0.024 | 6.995 ± 0.036 |
1.869 ± 0.018 | 7.322 ± 0.031 |
7.247 ± 0.043 | 9.27 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.305 ± 0.015 | 5.814 ± 0.029 |
3.7 ± 0.023 | 3.333 ± 0.017 |
3.74 ± 0.021 | 6.475 ± 0.028 |
5.675 ± 0.033 | 6.234 ± 0.024 |
1.273 ± 0.015 | 3.998 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
