
Bacterioplanes sanyensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Bacterioplanes
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
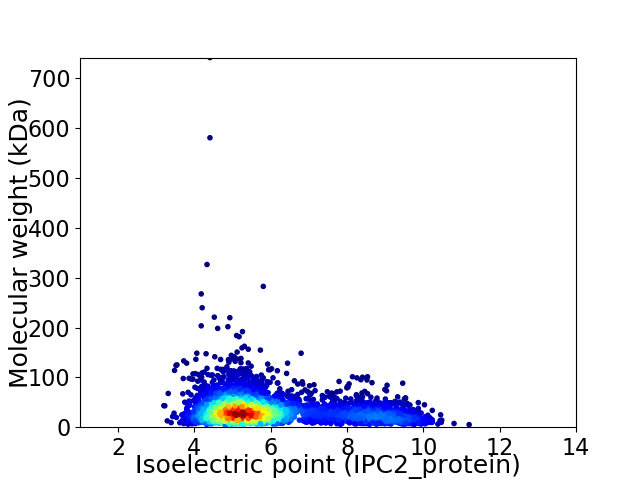
Virtual 2D-PAGE plot for 3784 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A222FHF0|A0A222FHF0_9GAMM N-acetyltransferase domain-containing protein OS=Bacterioplanes sanyensis OX=1249553 GN=CHH28_04085 PE=4 SV=1
MM1 pKa = 7.61ASSAQAQFIINEE13 pKa = 4.21LDD15 pKa = 3.27ADD17 pKa = 3.93QAGTDD22 pKa = 3.22SAEE25 pKa = 4.33FIEE28 pKa = 6.05LYY30 pKa = 10.76DD31 pKa = 4.39GGLGNQPLDD40 pKa = 3.91GYY42 pKa = 11.15SLVLYY47 pKa = 9.85NGSGSKK53 pKa = 10.15SYY55 pKa = 11.47NSVDD59 pKa = 3.27LSGYY63 pKa = 7.4STNADD68 pKa = 3.11GFFVVCGDD76 pKa = 3.62TAAVANCMLDD86 pKa = 2.57IGTNTNLIQNGADD99 pKa = 3.17AAALYY104 pKa = 9.72ARR106 pKa = 11.84PASDD110 pKa = 4.18FPSNTPVDD118 pKa = 3.91VNGVVDD124 pKa = 4.15ALVYY128 pKa = 8.56DD129 pKa = 4.51TNDD132 pKa = 3.1SDD134 pKa = 4.05ATALLALLNAGQPQVNEE151 pKa = 3.95AAGGDD156 pKa = 3.72KK157 pKa = 10.4DD158 pKa = 3.54AHH160 pKa = 6.29SNQRR164 pKa = 11.84CGDD167 pKa = 4.32GIARR171 pKa = 11.84NTDD174 pKa = 3.38GYY176 pKa = 10.17RR177 pKa = 11.84QQAPTPGAANQCDD190 pKa = 4.89GIDD193 pKa = 3.96PDD195 pKa = 4.78PGIGACGDD203 pKa = 3.73VSHH206 pKa = 7.33SDD208 pKa = 3.51YY209 pKa = 11.42QLISSIQGRR218 pKa = 11.84IEE220 pKa = 4.82DD221 pKa = 4.18SANDD225 pKa = 3.71ASPLNGQTVVVEE237 pKa = 4.26AVVTTDD243 pKa = 3.46LQGGQLANGDD253 pKa = 3.52NSFQYY258 pKa = 10.11SGYY261 pKa = 9.61WLQQLEE267 pKa = 4.38SEE269 pKa = 4.65YY270 pKa = 11.42DD271 pKa = 3.22NDD273 pKa = 3.78AQTSEE278 pKa = 4.71GIFVYY283 pKa = 10.05DD284 pKa = 3.89FRR286 pKa = 11.84SEE288 pKa = 4.08VSVGDD293 pKa = 3.68RR294 pKa = 11.84VRR296 pKa = 11.84LQAQVSEE303 pKa = 5.14FNQSTQLKK311 pKa = 8.75NVADD315 pKa = 4.29LVVCASGQSLPAPVALQLPVSDD337 pKa = 3.43KK338 pKa = 10.13TAWEE342 pKa = 4.09AVEE345 pKa = 4.54GMLVSSQQNLVVSDD359 pKa = 4.37LFGTGYY365 pKa = 11.26GFGNYY370 pKa = 8.93GQFVVSSRR378 pKa = 11.84LHH380 pKa = 4.92FQPTEE385 pKa = 3.91VEE387 pKa = 4.45LPGSAAAQALAAARR401 pKa = 11.84PLDD404 pKa = 4.14ALLIDD409 pKa = 4.74DD410 pKa = 5.42GVAKK414 pKa = 10.2SYY416 pKa = 10.3PAFIPFPDD424 pKa = 3.4EE425 pKa = 4.35SGFSASNPMRR435 pKa = 11.84IGYY438 pKa = 7.29QVPSVSGVMNEE449 pKa = 3.6FRR451 pKa = 11.84DD452 pKa = 4.05NYY454 pKa = 9.76TVIPNNIVIDD464 pKa = 3.93PVAARR469 pKa = 11.84TAAPQVALDD478 pKa = 3.75ADD480 pKa = 4.22LVVVGMNVLNYY491 pKa = 10.14FNGDD495 pKa = 3.22GQGGGFPTSRR505 pKa = 11.84GAPSAAAFEE514 pKa = 4.26MQTAKK519 pKa = 10.2IVAALQAMDD528 pKa = 4.03ADD530 pKa = 3.68IVGLMEE536 pKa = 5.2IEE538 pKa = 3.83NDD540 pKa = 4.14GYY542 pKa = 11.7GSDD545 pKa = 3.55SAIASLVNALNAVQAAGDD563 pKa = 3.82EE564 pKa = 4.31YY565 pKa = 10.83TYY567 pKa = 11.44VNPGRR572 pKa = 11.84NQVGTDD578 pKa = 4.18AIAVGLLYY586 pKa = 10.58RR587 pKa = 11.84PAKK590 pKa = 10.28VSLEE594 pKa = 4.06GASVILDD601 pKa = 3.66SSNSPLDD608 pKa = 3.53DD609 pKa = 3.18TGAPLFIDD617 pKa = 3.52NKK619 pKa = 10.15NRR621 pKa = 11.84PSLIQSFRR629 pKa = 11.84YY630 pKa = 9.5NDD632 pKa = 3.59TVFTLSVNHH641 pKa = 6.54LKK643 pKa = 10.89SKK645 pKa = 10.51GSPCGEE651 pKa = 4.1PNEE654 pKa = 4.54GEE656 pKa = 4.17NGVGSCNLTRR666 pKa = 11.84TTAAQALVQFLATNPTGVDD685 pKa = 3.0SDD687 pKa = 3.7ATLILGDD694 pKa = 4.17LNAYY698 pKa = 8.29SQEE701 pKa = 4.24DD702 pKa = 3.95PMQVFYY708 pKa = 11.12QGGFTNLKK716 pKa = 8.26YY717 pKa = 9.15TDD719 pKa = 4.2KK720 pKa = 11.11VSEE723 pKa = 4.09QQPFSYY729 pKa = 10.4SFSGFLGSLDD739 pKa = 3.66HH740 pKa = 7.12ALASPSLLNHH750 pKa = 5.64VVSVDD755 pKa = 2.75AWHH758 pKa = 6.76INSVEE763 pKa = 4.04APLMDD768 pKa = 4.34YY769 pKa = 9.71LTEE772 pKa = 4.85ANGQDD777 pKa = 3.78FDD779 pKa = 6.38SIDD782 pKa = 3.38NYY784 pKa = 11.14AAADD788 pKa = 3.95AYY790 pKa = 10.59RR791 pKa = 11.84SSDD794 pKa = 3.13HH795 pKa = 7.23DD796 pKa = 4.79PIVLGLSFAAANQAPQQSDD815 pKa = 4.21DD816 pKa = 3.65IADD819 pKa = 3.79VALEE823 pKa = 4.06QADD826 pKa = 3.88TLYY829 pKa = 11.12SIDD832 pKa = 3.59LSQYY836 pKa = 10.65FIDD839 pKa = 4.81ADD841 pKa = 3.94GDD843 pKa = 4.0ALSYY847 pKa = 10.9EE848 pKa = 4.74LDD850 pKa = 3.69GQQAGITLLATGEE863 pKa = 4.22LQVQLTAEE871 pKa = 4.24QLDD874 pKa = 4.2SLPLTLAFAVTDD886 pKa = 4.35GQASIAATITLLDD899 pKa = 3.59QRR901 pKa = 11.84PAQSPWQRR909 pKa = 11.84WWQWLQDD916 pKa = 3.18WWKK919 pKa = 10.66SLWDD923 pKa = 3.52
MM1 pKa = 7.61ASSAQAQFIINEE13 pKa = 4.21LDD15 pKa = 3.27ADD17 pKa = 3.93QAGTDD22 pKa = 3.22SAEE25 pKa = 4.33FIEE28 pKa = 6.05LYY30 pKa = 10.76DD31 pKa = 4.39GGLGNQPLDD40 pKa = 3.91GYY42 pKa = 11.15SLVLYY47 pKa = 9.85NGSGSKK53 pKa = 10.15SYY55 pKa = 11.47NSVDD59 pKa = 3.27LSGYY63 pKa = 7.4STNADD68 pKa = 3.11GFFVVCGDD76 pKa = 3.62TAAVANCMLDD86 pKa = 2.57IGTNTNLIQNGADD99 pKa = 3.17AAALYY104 pKa = 9.72ARR106 pKa = 11.84PASDD110 pKa = 4.18FPSNTPVDD118 pKa = 3.91VNGVVDD124 pKa = 4.15ALVYY128 pKa = 8.56DD129 pKa = 4.51TNDD132 pKa = 3.1SDD134 pKa = 4.05ATALLALLNAGQPQVNEE151 pKa = 3.95AAGGDD156 pKa = 3.72KK157 pKa = 10.4DD158 pKa = 3.54AHH160 pKa = 6.29SNQRR164 pKa = 11.84CGDD167 pKa = 4.32GIARR171 pKa = 11.84NTDD174 pKa = 3.38GYY176 pKa = 10.17RR177 pKa = 11.84QQAPTPGAANQCDD190 pKa = 4.89GIDD193 pKa = 3.96PDD195 pKa = 4.78PGIGACGDD203 pKa = 3.73VSHH206 pKa = 7.33SDD208 pKa = 3.51YY209 pKa = 11.42QLISSIQGRR218 pKa = 11.84IEE220 pKa = 4.82DD221 pKa = 4.18SANDD225 pKa = 3.71ASPLNGQTVVVEE237 pKa = 4.26AVVTTDD243 pKa = 3.46LQGGQLANGDD253 pKa = 3.52NSFQYY258 pKa = 10.11SGYY261 pKa = 9.61WLQQLEE267 pKa = 4.38SEE269 pKa = 4.65YY270 pKa = 11.42DD271 pKa = 3.22NDD273 pKa = 3.78AQTSEE278 pKa = 4.71GIFVYY283 pKa = 10.05DD284 pKa = 3.89FRR286 pKa = 11.84SEE288 pKa = 4.08VSVGDD293 pKa = 3.68RR294 pKa = 11.84VRR296 pKa = 11.84LQAQVSEE303 pKa = 5.14FNQSTQLKK311 pKa = 8.75NVADD315 pKa = 4.29LVVCASGQSLPAPVALQLPVSDD337 pKa = 3.43KK338 pKa = 10.13TAWEE342 pKa = 4.09AVEE345 pKa = 4.54GMLVSSQQNLVVSDD359 pKa = 4.37LFGTGYY365 pKa = 11.26GFGNYY370 pKa = 8.93GQFVVSSRR378 pKa = 11.84LHH380 pKa = 4.92FQPTEE385 pKa = 3.91VEE387 pKa = 4.45LPGSAAAQALAAARR401 pKa = 11.84PLDD404 pKa = 4.14ALLIDD409 pKa = 4.74DD410 pKa = 5.42GVAKK414 pKa = 10.2SYY416 pKa = 10.3PAFIPFPDD424 pKa = 3.4EE425 pKa = 4.35SGFSASNPMRR435 pKa = 11.84IGYY438 pKa = 7.29QVPSVSGVMNEE449 pKa = 3.6FRR451 pKa = 11.84DD452 pKa = 4.05NYY454 pKa = 9.76TVIPNNIVIDD464 pKa = 3.93PVAARR469 pKa = 11.84TAAPQVALDD478 pKa = 3.75ADD480 pKa = 4.22LVVVGMNVLNYY491 pKa = 10.14FNGDD495 pKa = 3.22GQGGGFPTSRR505 pKa = 11.84GAPSAAAFEE514 pKa = 4.26MQTAKK519 pKa = 10.2IVAALQAMDD528 pKa = 4.03ADD530 pKa = 3.68IVGLMEE536 pKa = 5.2IEE538 pKa = 3.83NDD540 pKa = 4.14GYY542 pKa = 11.7GSDD545 pKa = 3.55SAIASLVNALNAVQAAGDD563 pKa = 3.82EE564 pKa = 4.31YY565 pKa = 10.83TYY567 pKa = 11.44VNPGRR572 pKa = 11.84NQVGTDD578 pKa = 4.18AIAVGLLYY586 pKa = 10.58RR587 pKa = 11.84PAKK590 pKa = 10.28VSLEE594 pKa = 4.06GASVILDD601 pKa = 3.66SSNSPLDD608 pKa = 3.53DD609 pKa = 3.18TGAPLFIDD617 pKa = 3.52NKK619 pKa = 10.15NRR621 pKa = 11.84PSLIQSFRR629 pKa = 11.84YY630 pKa = 9.5NDD632 pKa = 3.59TVFTLSVNHH641 pKa = 6.54LKK643 pKa = 10.89SKK645 pKa = 10.51GSPCGEE651 pKa = 4.1PNEE654 pKa = 4.54GEE656 pKa = 4.17NGVGSCNLTRR666 pKa = 11.84TTAAQALVQFLATNPTGVDD685 pKa = 3.0SDD687 pKa = 3.7ATLILGDD694 pKa = 4.17LNAYY698 pKa = 8.29SQEE701 pKa = 4.24DD702 pKa = 3.95PMQVFYY708 pKa = 11.12QGGFTNLKK716 pKa = 8.26YY717 pKa = 9.15TDD719 pKa = 4.2KK720 pKa = 11.11VSEE723 pKa = 4.09QQPFSYY729 pKa = 10.4SFSGFLGSLDD739 pKa = 3.66HH740 pKa = 7.12ALASPSLLNHH750 pKa = 5.64VVSVDD755 pKa = 2.75AWHH758 pKa = 6.76INSVEE763 pKa = 4.04APLMDD768 pKa = 4.34YY769 pKa = 9.71LTEE772 pKa = 4.85ANGQDD777 pKa = 3.78FDD779 pKa = 6.38SIDD782 pKa = 3.38NYY784 pKa = 11.14AAADD788 pKa = 3.95AYY790 pKa = 10.59RR791 pKa = 11.84SSDD794 pKa = 3.13HH795 pKa = 7.23DD796 pKa = 4.79PIVLGLSFAAANQAPQQSDD815 pKa = 4.21DD816 pKa = 3.65IADD819 pKa = 3.79VALEE823 pKa = 4.06QADD826 pKa = 3.88TLYY829 pKa = 11.12SIDD832 pKa = 3.59LSQYY836 pKa = 10.65FIDD839 pKa = 4.81ADD841 pKa = 3.94GDD843 pKa = 4.0ALSYY847 pKa = 10.9EE848 pKa = 4.74LDD850 pKa = 3.69GQQAGITLLATGEE863 pKa = 4.22LQVQLTAEE871 pKa = 4.24QLDD874 pKa = 4.2SLPLTLAFAVTDD886 pKa = 4.35GQASIAATITLLDD899 pKa = 3.59QRR901 pKa = 11.84PAQSPWQRR909 pKa = 11.84WWQWLQDD916 pKa = 3.18WWKK919 pKa = 10.66SLWDD923 pKa = 3.52
Molecular weight: 97.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A222FFY9|A0A222FFY9_9GAMM Trk system potassium uptake protein OS=Bacterioplanes sanyensis OX=1249553 GN=CHH28_04520 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84HH40 pKa = 4.64VLSAA44 pKa = 3.81
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84HH40 pKa = 4.64VLSAA44 pKa = 3.81
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1254103 |
37 |
6764 |
331.4 |
36.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.042 ± 0.04 | 1.009 ± 0.017 |
5.999 ± 0.034 | 5.74 ± 0.038 |
3.55 ± 0.023 | 7.002 ± 0.04 |
2.541 ± 0.022 | 5.037 ± 0.028 |
3.361 ± 0.032 | 10.993 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.497 ± 0.022 | 3.434 ± 0.029 |
4.27 ± 0.025 | 6.151 ± 0.046 |
5.75 ± 0.036 | 6.419 ± 0.04 |
4.981 ± 0.031 | 6.874 ± 0.038 |
1.564 ± 0.02 | 2.785 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
