
Salinibacterium xinjiangense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Salinibacterium
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
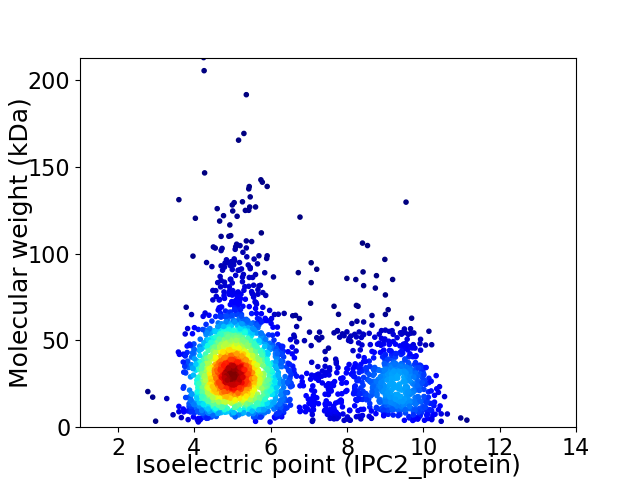
Virtual 2D-PAGE plot for 2810 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2C8YSW6|A0A2C8YSW6_9MICO Fur family transcriptional regulator ferric uptake regulator OS=Salinibacterium xinjiangense OX=386302 GN=SAMN06296378_0608 PE=3 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84ITRR5 pKa = 11.84TRR7 pKa = 11.84AIIVTGLVAGSLALTACSSPAAEE30 pKa = 4.6EE31 pKa = 4.04PASPSAAGAIDD42 pKa = 4.47LAAAGCPADD51 pKa = 5.46IKK53 pKa = 10.94IQTDD57 pKa = 3.37WNPEE61 pKa = 3.83AEE63 pKa = 4.3HH64 pKa = 5.63GHH66 pKa = 6.89LYY68 pKa = 10.94GLLGSDD74 pKa = 3.59YY75 pKa = 11.26TVDD78 pKa = 3.57TTNKK82 pKa = 8.59TVTGPLVVGGEE93 pKa = 4.09PTGVNVTILAGGPAVGFSQPNAQLYY118 pKa = 10.32SDD120 pKa = 5.2DD121 pKa = 5.03SIFMAYY127 pKa = 10.13VGTDD131 pKa = 3.07EE132 pKa = 6.01AIAHH136 pKa = 5.86SVDD139 pKa = 3.77LPTVGVFQPLEE150 pKa = 4.42KK151 pKa = 10.35DD152 pKa = 3.25PQMIMWDD159 pKa = 3.49PATYY163 pKa = 10.24PDD165 pKa = 3.93VEE167 pKa = 4.84SIADD171 pKa = 3.93LGKK174 pKa = 8.01TDD176 pKa = 3.1APIRR180 pKa = 11.84VFPGGVYY187 pKa = 9.07IDD189 pKa = 4.05YY190 pKa = 10.83FIGTGVLSADD200 pKa = 4.36QIDD203 pKa = 3.7ATYY206 pKa = 10.43DD207 pKa = 3.14GSPAVFVSEE216 pKa = 4.33GGKK219 pKa = 7.75TAQQGFASAEE229 pKa = 3.73PYY231 pKa = 9.81IYY233 pKa = 10.56KK234 pKa = 10.48NEE236 pKa = 3.83VAEE239 pKa = 4.05WGKK242 pKa = 10.11DD243 pKa = 3.28VKK245 pKa = 11.08YY246 pKa = 10.84QLVNDD251 pKa = 4.21AGFPKK256 pKa = 9.1YY257 pKa = 10.72AAMVSVRR264 pKa = 11.84PDD266 pKa = 3.27DD267 pKa = 3.49VTAYY271 pKa = 10.44ADD273 pKa = 3.87CLTALVPVMQQAEE286 pKa = 3.79IDD288 pKa = 4.26YY289 pKa = 11.16YY290 pKa = 11.47DD291 pKa = 4.03GTTLDD296 pKa = 3.63ATNALILDD304 pKa = 4.49LVDD307 pKa = 4.02QYY309 pKa = 11.75DD310 pKa = 3.85TGWVYY315 pKa = 10.99SQGVADD321 pKa = 4.24YY322 pKa = 10.97SVKK325 pKa = 9.49TQLADD330 pKa = 4.18EE331 pKa = 4.59IVSNGPDD338 pKa = 3.05ATLGNFDD345 pKa = 4.22DD346 pKa = 4.61ARR348 pKa = 11.84VAEE351 pKa = 4.6LFDD354 pKa = 3.46IVSPIFNKK362 pKa = 9.92QDD364 pKa = 2.76ITIADD369 pKa = 3.87GLAPTDD375 pKa = 3.98LYY377 pKa = 11.22TNEE380 pKa = 4.24FVDD383 pKa = 3.69MSLGLKK389 pKa = 9.76
MM1 pKa = 8.02RR2 pKa = 11.84ITRR5 pKa = 11.84TRR7 pKa = 11.84AIIVTGLVAGSLALTACSSPAAEE30 pKa = 4.6EE31 pKa = 4.04PASPSAAGAIDD42 pKa = 4.47LAAAGCPADD51 pKa = 5.46IKK53 pKa = 10.94IQTDD57 pKa = 3.37WNPEE61 pKa = 3.83AEE63 pKa = 4.3HH64 pKa = 5.63GHH66 pKa = 6.89LYY68 pKa = 10.94GLLGSDD74 pKa = 3.59YY75 pKa = 11.26TVDD78 pKa = 3.57TTNKK82 pKa = 8.59TVTGPLVVGGEE93 pKa = 4.09PTGVNVTILAGGPAVGFSQPNAQLYY118 pKa = 10.32SDD120 pKa = 5.2DD121 pKa = 5.03SIFMAYY127 pKa = 10.13VGTDD131 pKa = 3.07EE132 pKa = 6.01AIAHH136 pKa = 5.86SVDD139 pKa = 3.77LPTVGVFQPLEE150 pKa = 4.42KK151 pKa = 10.35DD152 pKa = 3.25PQMIMWDD159 pKa = 3.49PATYY163 pKa = 10.24PDD165 pKa = 3.93VEE167 pKa = 4.84SIADD171 pKa = 3.93LGKK174 pKa = 8.01TDD176 pKa = 3.1APIRR180 pKa = 11.84VFPGGVYY187 pKa = 9.07IDD189 pKa = 4.05YY190 pKa = 10.83FIGTGVLSADD200 pKa = 4.36QIDD203 pKa = 3.7ATYY206 pKa = 10.43DD207 pKa = 3.14GSPAVFVSEE216 pKa = 4.33GGKK219 pKa = 7.75TAQQGFASAEE229 pKa = 3.73PYY231 pKa = 9.81IYY233 pKa = 10.56KK234 pKa = 10.48NEE236 pKa = 3.83VAEE239 pKa = 4.05WGKK242 pKa = 10.11DD243 pKa = 3.28VKK245 pKa = 11.08YY246 pKa = 10.84QLVNDD251 pKa = 4.21AGFPKK256 pKa = 9.1YY257 pKa = 10.72AAMVSVRR264 pKa = 11.84PDD266 pKa = 3.27DD267 pKa = 3.49VTAYY271 pKa = 10.44ADD273 pKa = 3.87CLTALVPVMQQAEE286 pKa = 3.79IDD288 pKa = 4.26YY289 pKa = 11.16YY290 pKa = 11.47DD291 pKa = 4.03GTTLDD296 pKa = 3.63ATNALILDD304 pKa = 4.49LVDD307 pKa = 4.02QYY309 pKa = 11.75DD310 pKa = 3.85TGWVYY315 pKa = 10.99SQGVADD321 pKa = 4.24YY322 pKa = 10.97SVKK325 pKa = 9.49TQLADD330 pKa = 4.18EE331 pKa = 4.59IVSNGPDD338 pKa = 3.05ATLGNFDD345 pKa = 4.22DD346 pKa = 4.61ARR348 pKa = 11.84VAEE351 pKa = 4.6LFDD354 pKa = 3.46IVSPIFNKK362 pKa = 9.92QDD364 pKa = 2.76ITIADD369 pKa = 3.87GLAPTDD375 pKa = 3.98LYY377 pKa = 11.22TNEE380 pKa = 4.24FVDD383 pKa = 3.69MSLGLKK389 pKa = 9.76
Molecular weight: 41.19 kDa
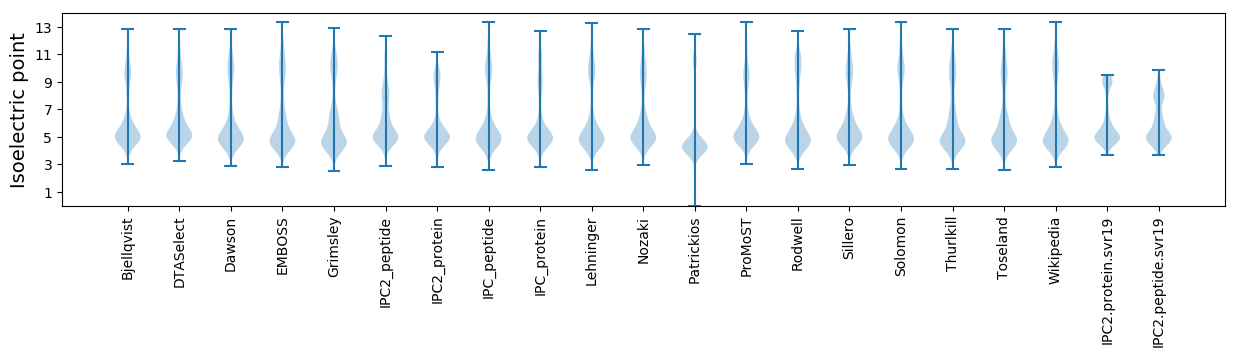
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2C9A2H8|A0A2C9A2H8_9MICO Peptidyl-prolyl cis-trans isomerase B (Cyclophilin B) OS=Salinibacterium xinjiangense OX=386302 GN=SAMN06296378_2753 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
898122 |
29 |
2018 |
319.6 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.546 ± 0.06 | 0.509 ± 0.011 |
5.997 ± 0.044 | 5.292 ± 0.048 |
3.383 ± 0.028 | 8.736 ± 0.042 |
1.895 ± 0.02 | 5.305 ± 0.035 |
2.349 ± 0.034 | 10.063 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.923 ± 0.019 | 2.443 ± 0.026 |
5.132 ± 0.03 | 2.881 ± 0.023 |
6.591 ± 0.045 | 6.457 ± 0.037 |
6.236 ± 0.043 | 8.811 ± 0.046 |
1.431 ± 0.02 | 2.018 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
