
Desulfobacterium autotrophicum (strain ATCC 43914 / DSM 3382 / HRM2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfobacterium; Desulfobacterium autotrophicum
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
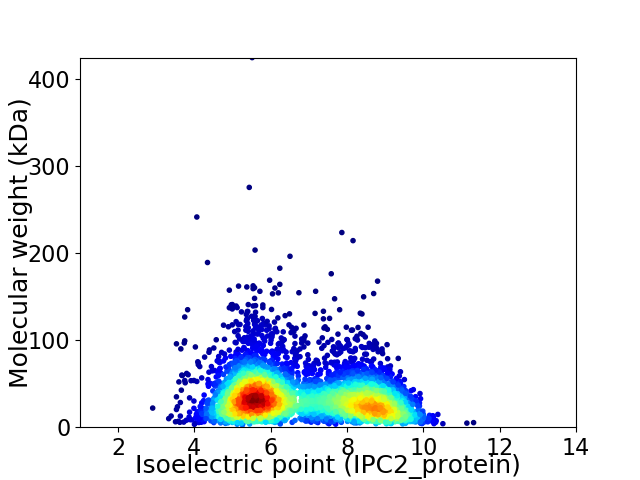
Virtual 2D-PAGE plot for 4846 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0QD89|C0QD89_DESAH Protein TonB OS=Desulfobacterium autotrophicum (strain ATCC 43914 / DSM 3382 / HRM2) OX=177437 GN=tonB PE=3 SV=1
MM1 pKa = 7.39SKK3 pKa = 9.73IEE5 pKa = 4.68RR6 pKa = 11.84ITHH9 pKa = 5.68KK10 pKa = 10.89SFMYY14 pKa = 9.12TPAIIVLLAAILAFSPLNATAMTFDD39 pKa = 3.8AGEE42 pKa = 4.05MGKK45 pKa = 8.43MEE47 pKa = 3.91VSGAIKK53 pKa = 10.41ASFGATQYY61 pKa = 11.79LDD63 pKa = 4.48FVDD66 pKa = 4.63NNSKK70 pKa = 11.06GDD72 pKa = 3.9DD73 pKa = 3.66ADD75 pKa = 4.14FDD77 pKa = 4.0ATRR80 pKa = 11.84EE81 pKa = 4.08LNLRR85 pKa = 11.84VDD87 pKa = 4.49WILNDD92 pKa = 3.42NLKK95 pKa = 10.87GVVSFQIGEE104 pKa = 4.31GSTGGYY110 pKa = 9.78FGSTDD115 pKa = 3.16AAVGGEE121 pKa = 3.99EE122 pKa = 4.92DD123 pKa = 3.87GDD125 pKa = 5.04LILEE129 pKa = 4.27LDD131 pKa = 4.15KK132 pKa = 11.39LYY134 pKa = 10.35IDD136 pKa = 3.82YY137 pKa = 7.79TTNGGVNFKK146 pKa = 10.67LGSQSFNLHH155 pKa = 6.12EE156 pKa = 4.19IAYY159 pKa = 9.17GSNIFYY165 pKa = 7.63EE166 pKa = 4.55TPPGVTMMAPLSEE179 pKa = 4.33TVSLEE184 pKa = 4.16VGWFRR189 pKa = 11.84MADD192 pKa = 3.37LMDD195 pKa = 5.09DD196 pKa = 4.35SEE198 pKa = 6.29SEE200 pKa = 4.15TDD202 pKa = 3.58DD203 pKa = 3.33QADD206 pKa = 4.34FIFAKK211 pKa = 9.82MPLSMGTVKK220 pKa = 9.17VTPWAAYY227 pKa = 10.5ANIQEE232 pKa = 4.66DD233 pKa = 4.66VIDD236 pKa = 3.8TDD238 pKa = 4.2EE239 pKa = 4.63DD240 pKa = 3.6NNYY243 pKa = 10.26YY244 pKa = 10.5KK245 pKa = 10.55YY246 pKa = 10.93AYY248 pKa = 9.78FDD250 pKa = 3.76YY251 pKa = 10.84PGLLTGANSAITNANPTDD269 pKa = 3.85NVNAYY274 pKa = 9.91YY275 pKa = 10.55LGASIQFSPNKK286 pKa = 9.53NLAVSASATYY296 pKa = 11.27GDD298 pKa = 5.1MDD300 pKa = 3.79WQTATVDD307 pKa = 3.47ASISGFFTDD316 pKa = 5.39LVVDD320 pKa = 3.71YY321 pKa = 10.56KK322 pKa = 9.77MASMTPEE329 pKa = 3.89FFVLYY334 pKa = 10.77GNGPDD339 pKa = 5.43ADD341 pKa = 4.68DD342 pKa = 4.82SDD344 pKa = 4.48IDD346 pKa = 3.88MMPVLIGGPTYY357 pKa = 9.7TSSYY361 pKa = 10.28FGGSRR366 pKa = 11.84FNDD369 pKa = 3.34NMFDD373 pKa = 4.34SYY375 pKa = 11.8DD376 pKa = 3.52STYY379 pKa = 9.48ATSMWAVGFKK389 pKa = 10.78LKK391 pKa = 10.49DD392 pKa = 3.05IRR394 pKa = 11.84TGEE397 pKa = 4.14RR398 pKa = 11.84LTHH401 pKa = 6.56EE402 pKa = 4.3FQIMYY407 pKa = 10.75AEE409 pKa = 4.52GTADD413 pKa = 4.13DD414 pKa = 5.75SIFQSPDD421 pKa = 3.74DD422 pKa = 3.53ILLNEE427 pKa = 4.75DD428 pKa = 3.39EE429 pKa = 5.33SLVEE433 pKa = 4.05VNFNSEE439 pKa = 4.19FQVMKK444 pKa = 10.21HH445 pKa = 5.5LVFATEE451 pKa = 4.28LGYY454 pKa = 8.73ITFDD458 pKa = 3.33EE459 pKa = 4.78DD460 pKa = 2.99SDD462 pKa = 4.31YY463 pKa = 11.81NEE465 pKa = 5.06EE466 pKa = 4.1INGDD470 pKa = 3.9VEE472 pKa = 4.98DD473 pKa = 4.32FWKK476 pKa = 10.29IACSIEE482 pKa = 4.07LSFF485 pKa = 5.66
MM1 pKa = 7.39SKK3 pKa = 9.73IEE5 pKa = 4.68RR6 pKa = 11.84ITHH9 pKa = 5.68KK10 pKa = 10.89SFMYY14 pKa = 9.12TPAIIVLLAAILAFSPLNATAMTFDD39 pKa = 3.8AGEE42 pKa = 4.05MGKK45 pKa = 8.43MEE47 pKa = 3.91VSGAIKK53 pKa = 10.41ASFGATQYY61 pKa = 11.79LDD63 pKa = 4.48FVDD66 pKa = 4.63NNSKK70 pKa = 11.06GDD72 pKa = 3.9DD73 pKa = 3.66ADD75 pKa = 4.14FDD77 pKa = 4.0ATRR80 pKa = 11.84EE81 pKa = 4.08LNLRR85 pKa = 11.84VDD87 pKa = 4.49WILNDD92 pKa = 3.42NLKK95 pKa = 10.87GVVSFQIGEE104 pKa = 4.31GSTGGYY110 pKa = 9.78FGSTDD115 pKa = 3.16AAVGGEE121 pKa = 3.99EE122 pKa = 4.92DD123 pKa = 3.87GDD125 pKa = 5.04LILEE129 pKa = 4.27LDD131 pKa = 4.15KK132 pKa = 11.39LYY134 pKa = 10.35IDD136 pKa = 3.82YY137 pKa = 7.79TTNGGVNFKK146 pKa = 10.67LGSQSFNLHH155 pKa = 6.12EE156 pKa = 4.19IAYY159 pKa = 9.17GSNIFYY165 pKa = 7.63EE166 pKa = 4.55TPPGVTMMAPLSEE179 pKa = 4.33TVSLEE184 pKa = 4.16VGWFRR189 pKa = 11.84MADD192 pKa = 3.37LMDD195 pKa = 5.09DD196 pKa = 4.35SEE198 pKa = 6.29SEE200 pKa = 4.15TDD202 pKa = 3.58DD203 pKa = 3.33QADD206 pKa = 4.34FIFAKK211 pKa = 9.82MPLSMGTVKK220 pKa = 9.17VTPWAAYY227 pKa = 10.5ANIQEE232 pKa = 4.66DD233 pKa = 4.66VIDD236 pKa = 3.8TDD238 pKa = 4.2EE239 pKa = 4.63DD240 pKa = 3.6NNYY243 pKa = 10.26YY244 pKa = 10.5KK245 pKa = 10.55YY246 pKa = 10.93AYY248 pKa = 9.78FDD250 pKa = 3.76YY251 pKa = 10.84PGLLTGANSAITNANPTDD269 pKa = 3.85NVNAYY274 pKa = 9.91YY275 pKa = 10.55LGASIQFSPNKK286 pKa = 9.53NLAVSASATYY296 pKa = 11.27GDD298 pKa = 5.1MDD300 pKa = 3.79WQTATVDD307 pKa = 3.47ASISGFFTDD316 pKa = 5.39LVVDD320 pKa = 3.71YY321 pKa = 10.56KK322 pKa = 9.77MASMTPEE329 pKa = 3.89FFVLYY334 pKa = 10.77GNGPDD339 pKa = 5.43ADD341 pKa = 4.68DD342 pKa = 4.82SDD344 pKa = 4.48IDD346 pKa = 3.88MMPVLIGGPTYY357 pKa = 9.7TSSYY361 pKa = 10.28FGGSRR366 pKa = 11.84FNDD369 pKa = 3.34NMFDD373 pKa = 4.34SYY375 pKa = 11.8DD376 pKa = 3.52STYY379 pKa = 9.48ATSMWAVGFKK389 pKa = 10.78LKK391 pKa = 10.49DD392 pKa = 3.05IRR394 pKa = 11.84TGEE397 pKa = 4.14RR398 pKa = 11.84LTHH401 pKa = 6.56EE402 pKa = 4.3FQIMYY407 pKa = 10.75AEE409 pKa = 4.52GTADD413 pKa = 4.13DD414 pKa = 5.75SIFQSPDD421 pKa = 3.74DD422 pKa = 3.53ILLNEE427 pKa = 4.75DD428 pKa = 3.39EE429 pKa = 5.33SLVEE433 pKa = 4.05VNFNSEE439 pKa = 4.19FQVMKK444 pKa = 10.21HH445 pKa = 5.5LVFATEE451 pKa = 4.28LGYY454 pKa = 8.73ITFDD458 pKa = 3.33EE459 pKa = 4.78DD460 pKa = 2.99SDD462 pKa = 4.31YY463 pKa = 11.81NEE465 pKa = 5.06EE466 pKa = 4.1INGDD470 pKa = 3.9VEE472 pKa = 4.98DD473 pKa = 4.32FWKK476 pKa = 10.29IACSIEE482 pKa = 4.07LSFF485 pKa = 5.66
Molecular weight: 53.58 kDa
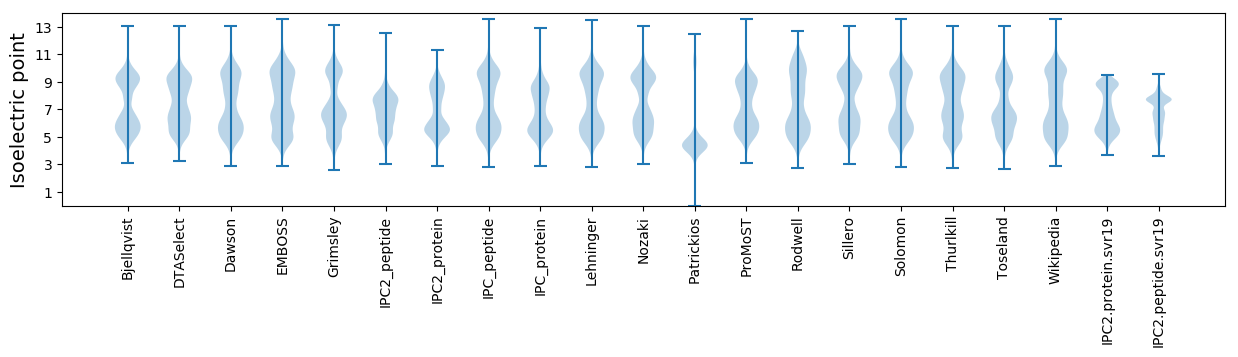
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0QJ31|C0QJ31_DESAH Fe-S oxidoreductase (Iron-sulfur cluster-binding domain protein) OS=Desulfobacterium autotrophicum (strain ATCC 43914 / DSM 3382 / HRM2) OX=177437 GN=HRM2_07070 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.19RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.67GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTAAGRR28 pKa = 11.84RR29 pKa = 11.84IVNSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.2KK41 pKa = 9.68LTAA44 pKa = 4.27
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.19RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.67GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTAAGRR28 pKa = 11.84RR29 pKa = 11.84IVNSRR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.2KK41 pKa = 9.68LTAA44 pKa = 4.27
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1634900 |
22 |
3859 |
337.4 |
37.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.026 ± 0.035 | 1.333 ± 0.014 |
5.537 ± 0.028 | 5.967 ± 0.033 |
4.625 ± 0.027 | 7.411 ± 0.033 |
2.069 ± 0.015 | 7.093 ± 0.028 |
6.0 ± 0.032 | 9.94 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.922 ± 0.016 | 3.903 ± 0.02 |
4.392 ± 0.023 | 3.449 ± 0.022 |
5.207 ± 0.024 | 5.862 ± 0.025 |
5.609 ± 0.027 | 6.814 ± 0.031 |
1.059 ± 0.013 | 2.78 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
