
Beihai picorna-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
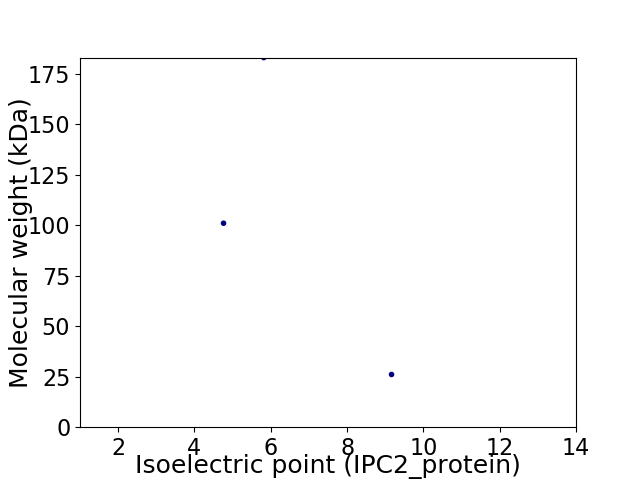
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
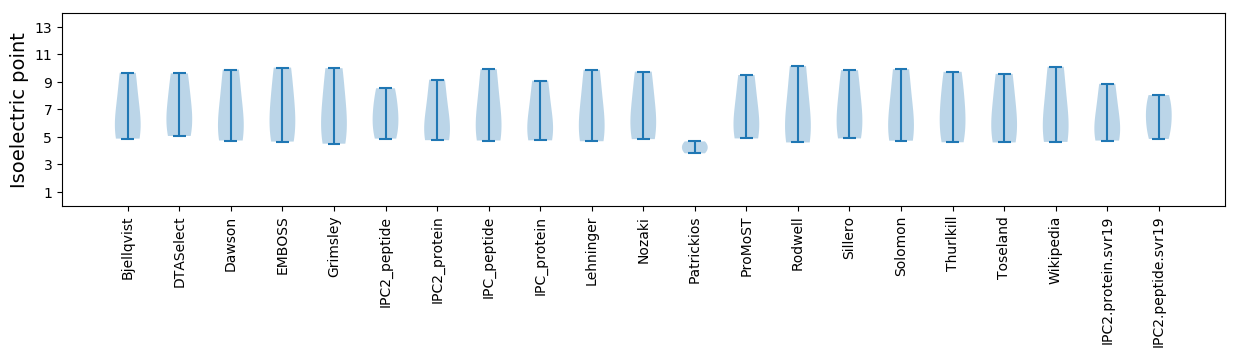
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHJ6|A0A1L3KHJ6_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 7 OX=1922616 PE=4 SV=1
MM1 pKa = 7.68PVLDD5 pKa = 5.6SIPEE9 pKa = 4.04NDD11 pKa = 5.7DD12 pKa = 3.62VLQCDD17 pKa = 3.44TRR19 pKa = 11.84LIKK22 pKa = 10.55PEE24 pKa = 4.25LFRR27 pKa = 11.84CATSGFTPQSGEE39 pKa = 3.51VDD41 pKa = 3.43QTVTQAPEE49 pKa = 4.06GTVEE53 pKa = 4.28QIVAFDD59 pKa = 3.79DD60 pKa = 5.05QEE62 pKa = 4.89AGWMKK67 pKa = 10.57DD68 pKa = 2.72ASGQFDD74 pKa = 3.57STMDD78 pKa = 3.9LVDD81 pKa = 5.7KK82 pKa = 11.21SDD84 pKa = 3.58TDD86 pKa = 3.59LGAFLGRR93 pKa = 11.84PVKK96 pKa = 10.15IDD98 pKa = 3.17TQAWVVGQPLYY109 pKa = 10.79KK110 pKa = 10.37QINPWEE116 pKa = 4.19LFVTHH121 pKa = 7.16PAMANKK127 pKa = 9.49LAHH130 pKa = 6.09YY131 pKa = 8.65EE132 pKa = 3.91LLRR135 pKa = 11.84GNLNVKK141 pKa = 9.33VVISGTGFHH150 pKa = 6.78YY151 pKa = 10.7GRR153 pKa = 11.84TLVSYY158 pKa = 10.89NPLNGYY164 pKa = 10.17DD165 pKa = 3.64EE166 pKa = 5.71ASVTRR171 pKa = 11.84NFIDD175 pKa = 3.21QDD177 pKa = 4.26LIAASQRR184 pKa = 11.84PCIFLNPTDD193 pKa = 3.78NSGGQMKK200 pKa = 10.7LPFFFLQNYY209 pKa = 6.72MSLSKK214 pKa = 10.93GDD216 pKa = 3.68YY217 pKa = 10.16RR218 pKa = 11.84DD219 pKa = 3.69MGVLDD224 pKa = 3.94YY225 pKa = 11.4KK226 pKa = 11.39SFGNLLHH233 pKa = 7.16ANGGDD238 pKa = 3.41DD239 pKa = 3.46PVYY242 pKa = 8.81ITTYY246 pKa = 10.61AWMTEE251 pKa = 4.12TSLTMPTSITSTITLQNEE269 pKa = 4.39TVDD272 pKa = 3.67PDD274 pKa = 4.04VFPLGPPYY282 pKa = 10.79GVGNGAEE289 pKa = 4.24NGVGPPFSPQSGFQPQAGGKK309 pKa = 9.46PNMKK313 pKa = 9.92KK314 pKa = 10.47SNKK317 pKa = 9.38LQSKK321 pKa = 9.87GNARR325 pKa = 11.84GKK327 pKa = 10.69AKK329 pKa = 10.76LNTGDD334 pKa = 4.34EE335 pKa = 4.37YY336 pKa = 11.47GQGIISAPASAVAKK350 pKa = 9.79AAGMLTSIPFIEE362 pKa = 5.27PYY364 pKa = 10.83ARR366 pKa = 11.84ATQMVASKK374 pKa = 9.83VGQVASIFGYY384 pKa = 9.93SRR386 pKa = 11.84PAVVSDD392 pKa = 4.31FLLQKK397 pKa = 10.27PSPTGNFANVDD408 pKa = 2.99ASDD411 pKa = 3.4AVNRR415 pKa = 11.84LTLDD419 pKa = 3.34SKK421 pKa = 11.18QEE423 pKa = 3.6ITIDD427 pKa = 3.46SRR429 pKa = 11.84TVGLDD434 pKa = 3.14GGDD437 pKa = 3.23QMAFDD442 pKa = 5.29NIQTRR447 pKa = 11.84EE448 pKa = 4.0SYY450 pKa = 9.07LTSFTMSSTDD460 pKa = 3.43APDD463 pKa = 4.79AILWNSYY470 pKa = 7.16VTPNLFGINNPEE482 pKa = 3.75LHH484 pKa = 5.63MTPMAMLAQYY494 pKa = 9.79FSAWQGTIKK503 pKa = 10.4FRR505 pKa = 11.84FQIVKK510 pKa = 10.78SNFHH514 pKa = 6.83KK515 pKa = 10.83GRR517 pKa = 11.84ILVRR521 pKa = 11.84YY522 pKa = 8.91DD523 pKa = 3.43PRR525 pKa = 11.84SHH527 pKa = 7.29SSDD530 pKa = 3.08VNYY533 pKa = 6.9TTNYY537 pKa = 9.87SRR539 pKa = 11.84VIDD542 pKa = 4.62LATEE546 pKa = 4.1DD547 pKa = 4.3DD548 pKa = 4.39FEE550 pKa = 6.08IEE552 pKa = 4.36IGWGQNVPFLGNQQANVSRR571 pKa = 11.84VYY573 pKa = 11.02YY574 pKa = 10.51GGTRR578 pKa = 11.84LTTDD582 pKa = 2.57SGGNHH587 pKa = 6.38NGVIEE592 pKa = 4.32VNVVNSLVAPAADD605 pKa = 3.22SDD607 pKa = 3.4IRR609 pKa = 11.84INVFVSAGDD618 pKa = 3.53DD619 pKa = 3.88MKK621 pKa = 10.47WGRR624 pKa = 11.84PNPDD628 pKa = 3.45KK629 pKa = 10.82IRR631 pKa = 11.84NLHH634 pKa = 5.83YY635 pKa = 10.45FPATAPPLNEE645 pKa = 4.21FQPQSGMGGDD655 pKa = 4.13LSGTTGDD662 pKa = 3.9SATDD666 pKa = 3.36RR667 pKa = 11.84PTEE670 pKa = 4.04ADD672 pKa = 3.21NVPSVGCPADD682 pKa = 4.64DD683 pKa = 3.79DD684 pKa = 5.15HH685 pKa = 8.17MMDD688 pKa = 3.32VFFGEE693 pKa = 4.46MPVSLRR699 pKa = 11.84EE700 pKa = 3.47IFRR703 pKa = 11.84RR704 pKa = 11.84YY705 pKa = 9.36VFEE708 pKa = 4.65KK709 pKa = 10.48YY710 pKa = 9.6FVPTAPTADD719 pKa = 3.62GALTTSNYY727 pKa = 9.76RR728 pKa = 11.84IKK730 pKa = 10.73SFPFHH735 pKa = 7.46SGDD738 pKa = 4.51DD739 pKa = 3.84PDD741 pKa = 5.26GLDD744 pKa = 3.42VKK746 pKa = 11.24SDD748 pKa = 3.31GLRR751 pKa = 11.84YY752 pKa = 9.83NQVQTSPIAFFAPCFAGWRR771 pKa = 11.84GSLRR775 pKa = 11.84HH776 pKa = 5.65KK777 pKa = 9.7WVFSGEE783 pKa = 4.3SLQSPLISNYY793 pKa = 10.05GFIAANGAQSTEE805 pKa = 3.8WTVTNDD811 pKa = 3.19LEE813 pKa = 4.58AEE815 pKa = 4.28KK816 pKa = 10.44FLSATFGKK824 pKa = 10.24FSNGGSAGTNLGVNNTIEE842 pKa = 4.29TEE844 pKa = 3.89MPYY847 pKa = 11.02YY848 pKa = 10.23NGNRR852 pKa = 11.84FSSSRR857 pKa = 11.84ILSADD862 pKa = 3.33NNSSYY867 pKa = 11.04SYY869 pKa = 10.26QVEE872 pKa = 4.87TIQSKK877 pKa = 9.68SVPSGPVPSVVTRR890 pKa = 11.84DD891 pKa = 2.88AFYY894 pKa = 11.03RR895 pKa = 11.84MCAAGEE901 pKa = 4.07DD902 pKa = 3.81FTLFFWTGVPIMYY915 pKa = 9.9NYY917 pKa = 10.35SRR919 pKa = 11.84TYY921 pKa = 9.9TSS923 pKa = 2.9
MM1 pKa = 7.68PVLDD5 pKa = 5.6SIPEE9 pKa = 4.04NDD11 pKa = 5.7DD12 pKa = 3.62VLQCDD17 pKa = 3.44TRR19 pKa = 11.84LIKK22 pKa = 10.55PEE24 pKa = 4.25LFRR27 pKa = 11.84CATSGFTPQSGEE39 pKa = 3.51VDD41 pKa = 3.43QTVTQAPEE49 pKa = 4.06GTVEE53 pKa = 4.28QIVAFDD59 pKa = 3.79DD60 pKa = 5.05QEE62 pKa = 4.89AGWMKK67 pKa = 10.57DD68 pKa = 2.72ASGQFDD74 pKa = 3.57STMDD78 pKa = 3.9LVDD81 pKa = 5.7KK82 pKa = 11.21SDD84 pKa = 3.58TDD86 pKa = 3.59LGAFLGRR93 pKa = 11.84PVKK96 pKa = 10.15IDD98 pKa = 3.17TQAWVVGQPLYY109 pKa = 10.79KK110 pKa = 10.37QINPWEE116 pKa = 4.19LFVTHH121 pKa = 7.16PAMANKK127 pKa = 9.49LAHH130 pKa = 6.09YY131 pKa = 8.65EE132 pKa = 3.91LLRR135 pKa = 11.84GNLNVKK141 pKa = 9.33VVISGTGFHH150 pKa = 6.78YY151 pKa = 10.7GRR153 pKa = 11.84TLVSYY158 pKa = 10.89NPLNGYY164 pKa = 10.17DD165 pKa = 3.64EE166 pKa = 5.71ASVTRR171 pKa = 11.84NFIDD175 pKa = 3.21QDD177 pKa = 4.26LIAASQRR184 pKa = 11.84PCIFLNPTDD193 pKa = 3.78NSGGQMKK200 pKa = 10.7LPFFFLQNYY209 pKa = 6.72MSLSKK214 pKa = 10.93GDD216 pKa = 3.68YY217 pKa = 10.16RR218 pKa = 11.84DD219 pKa = 3.69MGVLDD224 pKa = 3.94YY225 pKa = 11.4KK226 pKa = 11.39SFGNLLHH233 pKa = 7.16ANGGDD238 pKa = 3.41DD239 pKa = 3.46PVYY242 pKa = 8.81ITTYY246 pKa = 10.61AWMTEE251 pKa = 4.12TSLTMPTSITSTITLQNEE269 pKa = 4.39TVDD272 pKa = 3.67PDD274 pKa = 4.04VFPLGPPYY282 pKa = 10.79GVGNGAEE289 pKa = 4.24NGVGPPFSPQSGFQPQAGGKK309 pKa = 9.46PNMKK313 pKa = 9.92KK314 pKa = 10.47SNKK317 pKa = 9.38LQSKK321 pKa = 9.87GNARR325 pKa = 11.84GKK327 pKa = 10.69AKK329 pKa = 10.76LNTGDD334 pKa = 4.34EE335 pKa = 4.37YY336 pKa = 11.47GQGIISAPASAVAKK350 pKa = 9.79AAGMLTSIPFIEE362 pKa = 5.27PYY364 pKa = 10.83ARR366 pKa = 11.84ATQMVASKK374 pKa = 9.83VGQVASIFGYY384 pKa = 9.93SRR386 pKa = 11.84PAVVSDD392 pKa = 4.31FLLQKK397 pKa = 10.27PSPTGNFANVDD408 pKa = 2.99ASDD411 pKa = 3.4AVNRR415 pKa = 11.84LTLDD419 pKa = 3.34SKK421 pKa = 11.18QEE423 pKa = 3.6ITIDD427 pKa = 3.46SRR429 pKa = 11.84TVGLDD434 pKa = 3.14GGDD437 pKa = 3.23QMAFDD442 pKa = 5.29NIQTRR447 pKa = 11.84EE448 pKa = 4.0SYY450 pKa = 9.07LTSFTMSSTDD460 pKa = 3.43APDD463 pKa = 4.79AILWNSYY470 pKa = 7.16VTPNLFGINNPEE482 pKa = 3.75LHH484 pKa = 5.63MTPMAMLAQYY494 pKa = 9.79FSAWQGTIKK503 pKa = 10.4FRR505 pKa = 11.84FQIVKK510 pKa = 10.78SNFHH514 pKa = 6.83KK515 pKa = 10.83GRR517 pKa = 11.84ILVRR521 pKa = 11.84YY522 pKa = 8.91DD523 pKa = 3.43PRR525 pKa = 11.84SHH527 pKa = 7.29SSDD530 pKa = 3.08VNYY533 pKa = 6.9TTNYY537 pKa = 9.87SRR539 pKa = 11.84VIDD542 pKa = 4.62LATEE546 pKa = 4.1DD547 pKa = 4.3DD548 pKa = 4.39FEE550 pKa = 6.08IEE552 pKa = 4.36IGWGQNVPFLGNQQANVSRR571 pKa = 11.84VYY573 pKa = 11.02YY574 pKa = 10.51GGTRR578 pKa = 11.84LTTDD582 pKa = 2.57SGGNHH587 pKa = 6.38NGVIEE592 pKa = 4.32VNVVNSLVAPAADD605 pKa = 3.22SDD607 pKa = 3.4IRR609 pKa = 11.84INVFVSAGDD618 pKa = 3.53DD619 pKa = 3.88MKK621 pKa = 10.47WGRR624 pKa = 11.84PNPDD628 pKa = 3.45KK629 pKa = 10.82IRR631 pKa = 11.84NLHH634 pKa = 5.83YY635 pKa = 10.45FPATAPPLNEE645 pKa = 4.21FQPQSGMGGDD655 pKa = 4.13LSGTTGDD662 pKa = 3.9SATDD666 pKa = 3.36RR667 pKa = 11.84PTEE670 pKa = 4.04ADD672 pKa = 3.21NVPSVGCPADD682 pKa = 4.64DD683 pKa = 3.79DD684 pKa = 5.15HH685 pKa = 8.17MMDD688 pKa = 3.32VFFGEE693 pKa = 4.46MPVSLRR699 pKa = 11.84EE700 pKa = 3.47IFRR703 pKa = 11.84RR704 pKa = 11.84YY705 pKa = 9.36VFEE708 pKa = 4.65KK709 pKa = 10.48YY710 pKa = 9.6FVPTAPTADD719 pKa = 3.62GALTTSNYY727 pKa = 9.76RR728 pKa = 11.84IKK730 pKa = 10.73SFPFHH735 pKa = 7.46SGDD738 pKa = 4.51DD739 pKa = 3.84PDD741 pKa = 5.26GLDD744 pKa = 3.42VKK746 pKa = 11.24SDD748 pKa = 3.31GLRR751 pKa = 11.84YY752 pKa = 9.83NQVQTSPIAFFAPCFAGWRR771 pKa = 11.84GSLRR775 pKa = 11.84HH776 pKa = 5.65KK777 pKa = 9.7WVFSGEE783 pKa = 4.3SLQSPLISNYY793 pKa = 10.05GFIAANGAQSTEE805 pKa = 3.8WTVTNDD811 pKa = 3.19LEE813 pKa = 4.58AEE815 pKa = 4.28KK816 pKa = 10.44FLSATFGKK824 pKa = 10.24FSNGGSAGTNLGVNNTIEE842 pKa = 4.29TEE844 pKa = 3.89MPYY847 pKa = 11.02YY848 pKa = 10.23NGNRR852 pKa = 11.84FSSSRR857 pKa = 11.84ILSADD862 pKa = 3.33NNSSYY867 pKa = 11.04SYY869 pKa = 10.26QVEE872 pKa = 4.87TIQSKK877 pKa = 9.68SVPSGPVPSVVTRR890 pKa = 11.84DD891 pKa = 2.88AFYY894 pKa = 11.03RR895 pKa = 11.84MCAAGEE901 pKa = 4.07DD902 pKa = 3.81FTLFFWTGVPIMYY915 pKa = 9.9NYY917 pKa = 10.35SRR919 pKa = 11.84TYY921 pKa = 9.9TSS923 pKa = 2.9
Molecular weight: 101.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHM6|A0A1L3KHM6_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 7 OX=1922616 PE=4 SV=1
MM1 pKa = 7.09GQSPCTWANRR11 pKa = 11.84PNKK14 pKa = 8.87TLYY17 pKa = 10.62FVMFQINSKK26 pKa = 8.97TKK28 pKa = 10.52LDD30 pKa = 3.53VSSARR35 pKa = 11.84GRR37 pKa = 11.84KK38 pKa = 8.2SKK40 pKa = 10.39SYY42 pKa = 10.6RR43 pKa = 11.84GTLQGLRR50 pKa = 11.84IQQCLFVGAGWFLTSYY66 pKa = 10.01MPIRR70 pKa = 11.84EE71 pKa = 4.29CTTLSFEE78 pKa = 4.09GLSYY82 pKa = 11.0DD83 pKa = 3.45FQLFVPLFRR92 pKa = 11.84PTCDD96 pKa = 3.01FVVALYY102 pKa = 10.78VEE104 pKa = 4.56EE105 pKa = 4.42LADD108 pKa = 3.7VQSKK112 pKa = 9.22VVLQSDD118 pKa = 4.06APDD121 pKa = 3.27SFQYY125 pKa = 10.9LFSGKK130 pKa = 8.44SHH132 pKa = 7.06SLTKK136 pKa = 9.76MVPYY140 pKa = 9.84YY141 pKa = 11.15GKK143 pKa = 10.3GQRR146 pKa = 11.84ADD148 pKa = 3.32IIKK151 pKa = 9.66CDD153 pKa = 3.72EE154 pKa = 3.91MALVYY159 pKa = 10.31GINKK163 pKa = 9.73YY164 pKa = 10.07ILPKK168 pKa = 10.41FSFFRR173 pKa = 11.84FKK175 pKa = 10.03THH177 pKa = 5.98LCEE180 pKa = 4.1VCKK183 pKa = 11.01SNVNGRR189 pKa = 11.84VTYY192 pKa = 10.44GVLAYY197 pKa = 10.49HH198 pKa = 6.82KK199 pKa = 8.47TQDD202 pKa = 2.94WKK204 pKa = 11.02RR205 pKa = 11.84GNKK208 pKa = 9.87LGGLRR213 pKa = 11.84VYY215 pKa = 9.38TQGTLSATYY224 pKa = 10.26KK225 pKa = 10.55RR226 pKa = 11.84LRR228 pKa = 3.69
MM1 pKa = 7.09GQSPCTWANRR11 pKa = 11.84PNKK14 pKa = 8.87TLYY17 pKa = 10.62FVMFQINSKK26 pKa = 8.97TKK28 pKa = 10.52LDD30 pKa = 3.53VSSARR35 pKa = 11.84GRR37 pKa = 11.84KK38 pKa = 8.2SKK40 pKa = 10.39SYY42 pKa = 10.6RR43 pKa = 11.84GTLQGLRR50 pKa = 11.84IQQCLFVGAGWFLTSYY66 pKa = 10.01MPIRR70 pKa = 11.84EE71 pKa = 4.29CTTLSFEE78 pKa = 4.09GLSYY82 pKa = 11.0DD83 pKa = 3.45FQLFVPLFRR92 pKa = 11.84PTCDD96 pKa = 3.01FVVALYY102 pKa = 10.78VEE104 pKa = 4.56EE105 pKa = 4.42LADD108 pKa = 3.7VQSKK112 pKa = 9.22VVLQSDD118 pKa = 4.06APDD121 pKa = 3.27SFQYY125 pKa = 10.9LFSGKK130 pKa = 8.44SHH132 pKa = 7.06SLTKK136 pKa = 9.76MVPYY140 pKa = 9.84YY141 pKa = 11.15GKK143 pKa = 10.3GQRR146 pKa = 11.84ADD148 pKa = 3.32IIKK151 pKa = 9.66CDD153 pKa = 3.72EE154 pKa = 3.91MALVYY159 pKa = 10.31GINKK163 pKa = 9.73YY164 pKa = 10.07ILPKK168 pKa = 10.41FSFFRR173 pKa = 11.84FKK175 pKa = 10.03THH177 pKa = 5.98LCEE180 pKa = 4.1VCKK183 pKa = 11.01SNVNGRR189 pKa = 11.84VTYY192 pKa = 10.44GVLAYY197 pKa = 10.49HH198 pKa = 6.82KK199 pKa = 8.47TQDD202 pKa = 2.94WKK204 pKa = 11.02RR205 pKa = 11.84GNKK208 pKa = 9.87LGGLRR213 pKa = 11.84VYY215 pKa = 9.38TQGTLSATYY224 pKa = 10.26KK225 pKa = 10.55RR226 pKa = 11.84LRR228 pKa = 3.69
Molecular weight: 26.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2764 |
228 |
1613 |
921.3 |
103.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.368 ± 0.549 | 1.23 ± 0.477 |
6.766 ± 0.672 | 4.522 ± 0.681 |
6.006 ± 0.205 | 6.766 ± 0.931 |
1.7 ± 0.235 | 5.174 ± 0.686 |
5.644 ± 1.046 | 8.068 ± 0.864 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.075 ± 0.309 | 4.957 ± 0.672 |
4.848 ± 0.758 | 3.654 ± 0.567 |
4.667 ± 0.429 | 8.466 ± 0.153 |
5.861 ± 0.743 | 6.838 ± 0.15 |
1.447 ± 0.085 | 3.944 ± 0.532 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
