
Sulfurospirillum sp. UBA12182
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Sulfurospirillum; unclassified Sulfurospirillum
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
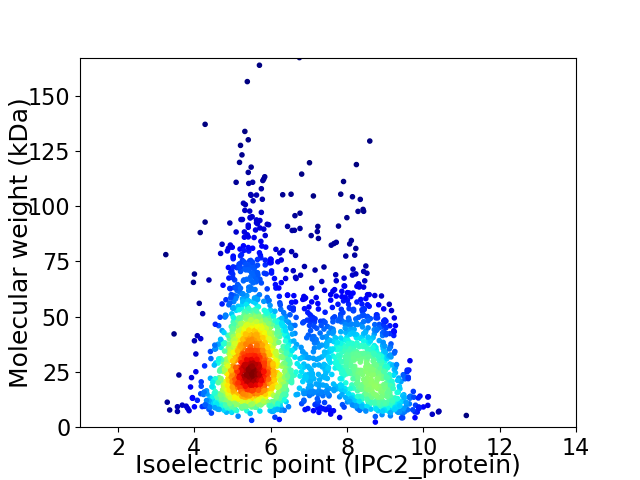
Virtual 2D-PAGE plot for 2326 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D3W5W6|A0A2D3W5W6_9PROT Ketol-acid reductoisomerase (NADP(+)) OS=Sulfurospirillum sp. UBA12182 OX=2015905 GN=ilvC PE=3 SV=1
MM1 pKa = 6.89NTSFYY6 pKa = 11.29NGISGIKK13 pKa = 9.1SHH15 pKa = 6.71QFGLDD20 pKa = 2.82VWADD24 pKa = 3.65NISNISTIGYY34 pKa = 8.71KK35 pKa = 10.0AANPEE40 pKa = 3.8FSTIFSTTLANSYY53 pKa = 11.22SDD55 pKa = 3.41PTSNNQGLGSRR66 pKa = 11.84AQATTLNMSQGIFQNTDD83 pKa = 2.46NVFDD87 pKa = 4.58LSIGGEE93 pKa = 4.17GWFGVQSLEE102 pKa = 3.77NQTYY106 pKa = 6.17YY107 pKa = 10.49TRR109 pKa = 11.84AGSFSMDD116 pKa = 2.9TDD118 pKa = 4.27GYY120 pKa = 11.44LVDD123 pKa = 5.15AYY125 pKa = 10.88GNYY128 pKa = 10.74LLGTLGGNVTPTSLSDD144 pKa = 3.87RR145 pKa = 11.84ILNEE149 pKa = 3.24FGTYY153 pKa = 10.15YY154 pKa = 10.86KK155 pKa = 10.74VGNTQEE161 pKa = 4.85LGTPYY166 pKa = 10.25SISALEE172 pKa = 5.19DD173 pKa = 2.97ITLSSVEE180 pKa = 4.21SQSKK184 pKa = 10.52INLPDD189 pKa = 3.1ILYY192 pKa = 9.28FPPEE196 pKa = 3.35ATTYY200 pKa = 10.94VKK202 pKa = 10.79YY203 pKa = 10.09QANLNPKK210 pKa = 9.57INIGSTKK217 pKa = 9.69IDD219 pKa = 4.97LDD221 pKa = 3.74TLDD224 pKa = 4.39IGTPAIDD231 pKa = 3.85TVNEE235 pKa = 4.1TLSISGTVSNTTALQNPQSGDD256 pKa = 3.6VVILTVTDD264 pKa = 3.72EE265 pKa = 4.53NGNSVDD271 pKa = 3.09ITAEE275 pKa = 4.05LTPLSSTDD283 pKa = 3.28STLVWSFSNSDD294 pKa = 3.08ISDD297 pKa = 4.23LDD299 pKa = 3.66TSTSLTITAQLQTEE313 pKa = 4.16QEE315 pKa = 4.24IPNVEE320 pKa = 4.16HH321 pKa = 6.45FTTEE325 pKa = 3.86IISPEE330 pKa = 4.09GEE332 pKa = 3.83KK333 pKa = 11.03DD334 pKa = 3.93FIDD337 pKa = 3.3MTFTKK342 pKa = 10.22RR343 pKa = 11.84VPQSALEE350 pKa = 4.36TTWDD354 pKa = 3.46AQIQILSFYY363 pKa = 9.62EE364 pKa = 4.19DD365 pKa = 4.05YY366 pKa = 11.15VIQTYY371 pKa = 10.43DD372 pKa = 3.19PSVTYY377 pKa = 10.63DD378 pKa = 3.22PTLYY382 pKa = 10.65NVNLEE387 pKa = 4.4TNQVTKK393 pKa = 10.35IYY395 pKa = 10.79DD396 pKa = 3.35PTLYY400 pKa = 10.71KK401 pKa = 10.38VDD403 pKa = 3.84LGTNKK408 pKa = 9.45VYY410 pKa = 10.66EE411 pKa = 5.04IIDD414 pKa = 3.86SQIGDD419 pKa = 3.59ATFAGAGEE427 pKa = 5.05LIASNMPTLSNGGTPLSINIGTPYY451 pKa = 10.73AEE453 pKa = 4.29EE454 pKa = 4.05NGVIISNGYY463 pKa = 10.4DD464 pKa = 3.17GMVSHH469 pKa = 7.34VDD471 pKa = 3.35LDD473 pKa = 3.54KK474 pKa = 11.45ARR476 pKa = 11.84YY477 pKa = 8.25SEE479 pKa = 3.9KK480 pKa = 10.62DD481 pKa = 3.35GYY483 pKa = 11.53VEE485 pKa = 4.64GLLKK489 pKa = 10.57AYY491 pKa = 10.29GMDD494 pKa = 3.08GRR496 pKa = 11.84GNVIAEE502 pKa = 4.09FTNGRR507 pKa = 11.84SAPIAKK513 pKa = 9.41IALYY517 pKa = 10.65HH518 pKa = 6.2FMNDD522 pKa = 2.92QGLEE526 pKa = 4.28KK527 pKa = 10.74VSSTLFSASANSGDD541 pKa = 4.12AIFYY545 pKa = 10.09TDD547 pKa = 4.14ANGNPILGSAIYY559 pKa = 10.34SNRR562 pKa = 11.84LEE564 pKa = 4.21GSNVSMATALTEE576 pKa = 4.51LIVMQKK582 pKa = 10.76AFDD585 pKa = 3.92ASAKK589 pKa = 10.62SITTSDD595 pKa = 3.52QMIQNAINMKK605 pKa = 10.24AA606 pKa = 3.04
MM1 pKa = 6.89NTSFYY6 pKa = 11.29NGISGIKK13 pKa = 9.1SHH15 pKa = 6.71QFGLDD20 pKa = 2.82VWADD24 pKa = 3.65NISNISTIGYY34 pKa = 8.71KK35 pKa = 10.0AANPEE40 pKa = 3.8FSTIFSTTLANSYY53 pKa = 11.22SDD55 pKa = 3.41PTSNNQGLGSRR66 pKa = 11.84AQATTLNMSQGIFQNTDD83 pKa = 2.46NVFDD87 pKa = 4.58LSIGGEE93 pKa = 4.17GWFGVQSLEE102 pKa = 3.77NQTYY106 pKa = 6.17YY107 pKa = 10.49TRR109 pKa = 11.84AGSFSMDD116 pKa = 2.9TDD118 pKa = 4.27GYY120 pKa = 11.44LVDD123 pKa = 5.15AYY125 pKa = 10.88GNYY128 pKa = 10.74LLGTLGGNVTPTSLSDD144 pKa = 3.87RR145 pKa = 11.84ILNEE149 pKa = 3.24FGTYY153 pKa = 10.15YY154 pKa = 10.86KK155 pKa = 10.74VGNTQEE161 pKa = 4.85LGTPYY166 pKa = 10.25SISALEE172 pKa = 5.19DD173 pKa = 2.97ITLSSVEE180 pKa = 4.21SQSKK184 pKa = 10.52INLPDD189 pKa = 3.1ILYY192 pKa = 9.28FPPEE196 pKa = 3.35ATTYY200 pKa = 10.94VKK202 pKa = 10.79YY203 pKa = 10.09QANLNPKK210 pKa = 9.57INIGSTKK217 pKa = 9.69IDD219 pKa = 4.97LDD221 pKa = 3.74TLDD224 pKa = 4.39IGTPAIDD231 pKa = 3.85TVNEE235 pKa = 4.1TLSISGTVSNTTALQNPQSGDD256 pKa = 3.6VVILTVTDD264 pKa = 3.72EE265 pKa = 4.53NGNSVDD271 pKa = 3.09ITAEE275 pKa = 4.05LTPLSSTDD283 pKa = 3.28STLVWSFSNSDD294 pKa = 3.08ISDD297 pKa = 4.23LDD299 pKa = 3.66TSTSLTITAQLQTEE313 pKa = 4.16QEE315 pKa = 4.24IPNVEE320 pKa = 4.16HH321 pKa = 6.45FTTEE325 pKa = 3.86IISPEE330 pKa = 4.09GEE332 pKa = 3.83KK333 pKa = 11.03DD334 pKa = 3.93FIDD337 pKa = 3.3MTFTKK342 pKa = 10.22RR343 pKa = 11.84VPQSALEE350 pKa = 4.36TTWDD354 pKa = 3.46AQIQILSFYY363 pKa = 9.62EE364 pKa = 4.19DD365 pKa = 4.05YY366 pKa = 11.15VIQTYY371 pKa = 10.43DD372 pKa = 3.19PSVTYY377 pKa = 10.63DD378 pKa = 3.22PTLYY382 pKa = 10.65NVNLEE387 pKa = 4.4TNQVTKK393 pKa = 10.35IYY395 pKa = 10.79DD396 pKa = 3.35PTLYY400 pKa = 10.71KK401 pKa = 10.38VDD403 pKa = 3.84LGTNKK408 pKa = 9.45VYY410 pKa = 10.66EE411 pKa = 5.04IIDD414 pKa = 3.86SQIGDD419 pKa = 3.59ATFAGAGEE427 pKa = 5.05LIASNMPTLSNGGTPLSINIGTPYY451 pKa = 10.73AEE453 pKa = 4.29EE454 pKa = 4.05NGVIISNGYY463 pKa = 10.4DD464 pKa = 3.17GMVSHH469 pKa = 7.34VDD471 pKa = 3.35LDD473 pKa = 3.54KK474 pKa = 11.45ARR476 pKa = 11.84YY477 pKa = 8.25SEE479 pKa = 3.9KK480 pKa = 10.62DD481 pKa = 3.35GYY483 pKa = 11.53VEE485 pKa = 4.64GLLKK489 pKa = 10.57AYY491 pKa = 10.29GMDD494 pKa = 3.08GRR496 pKa = 11.84GNVIAEE502 pKa = 4.09FTNGRR507 pKa = 11.84SAPIAKK513 pKa = 9.41IALYY517 pKa = 10.65HH518 pKa = 6.2FMNDD522 pKa = 2.92QGLEE526 pKa = 4.28KK527 pKa = 10.74VSSTLFSASANSGDD541 pKa = 4.12AIFYY545 pKa = 10.09TDD547 pKa = 4.14ANGNPILGSAIYY559 pKa = 10.34SNRR562 pKa = 11.84LEE564 pKa = 4.21GSNVSMATALTEE576 pKa = 4.51LIVMQKK582 pKa = 10.76AFDD585 pKa = 3.92ASAKK589 pKa = 10.62SITTSDD595 pKa = 3.52QMIQNAINMKK605 pKa = 10.24AA606 pKa = 3.04
Molecular weight: 65.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D3W5G9|A0A2D3W5G9_9PROT TonB-dependent receptor OS=Sulfurospirillum sp. UBA12182 OX=2015905 GN=CFH82_10390 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.63TKK25 pKa = 9.65NGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.68RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.31RR14 pKa = 11.84THH16 pKa = 5.88GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.63TKK25 pKa = 9.65NGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.76GRR39 pKa = 11.84KK40 pKa = 8.68RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
698452 |
20 |
1506 |
300.3 |
33.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.56 ± 0.048 | 1.016 ± 0.021 |
5.309 ± 0.036 | 7.507 ± 0.053 |
5.389 ± 0.044 | 5.854 ± 0.053 |
1.857 ± 0.022 | 8.508 ± 0.047 |
8.79 ± 0.053 | 10.23 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.446 ± 0.023 | 4.96 ± 0.045 |
2.97 ± 0.03 | 3.174 ± 0.027 |
3.301 ± 0.034 | 6.628 ± 0.041 |
4.722 ± 0.034 | 6.227 ± 0.042 |
0.726 ± 0.016 | 3.827 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
