
Dethiobacter alkaliphilus AHT 1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Syntrophomonadaceae; Dethiobacter; Dethiobacter alkaliphilus
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
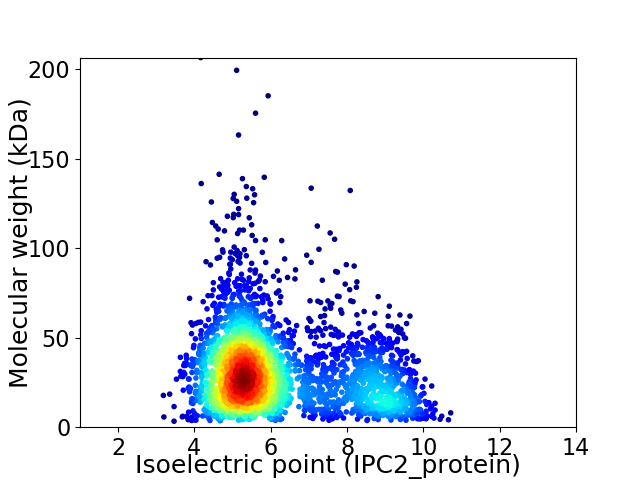
Virtual 2D-PAGE plot for 3162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0GEF8|C0GEF8_9FIRM Diguanylate cyclase and metal dependent phosphohydrolase OS=Dethiobacter alkaliphilus AHT 1 OX=555088 GN=DealDRAFT_0867 PE=4 SV=1
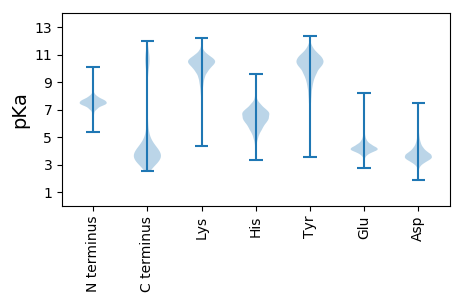
MM1 pKa = 7.28NMAKK5 pKa = 9.96PVSKK9 pKa = 9.91YY10 pKa = 7.29WRR12 pKa = 11.84VGLVLLVIGLLAIGTALGSVPPCEE36 pKa = 4.74GPLVDD41 pKa = 3.48PVEE44 pKa = 4.36YY45 pKa = 10.62NGNPEE50 pKa = 4.31EE51 pKa = 4.35IGTRR55 pKa = 11.84INDD58 pKa = 3.94PGSGYY63 pKa = 9.99IDD65 pKa = 3.52VEE67 pKa = 4.57FDD69 pKa = 3.67GVWYY73 pKa = 10.2KK74 pKa = 10.79VFYY77 pKa = 10.24EE78 pKa = 4.55VYY80 pKa = 10.58DD81 pKa = 3.59GGKK84 pKa = 7.18TLKK87 pKa = 10.04FWEE90 pKa = 4.46EE91 pKa = 3.45NGFPVVSKK99 pKa = 9.12VTVKK103 pKa = 10.77GGPNANIYY111 pKa = 10.26YY112 pKa = 9.79YY113 pKa = 10.94AADD116 pKa = 3.99PPEE119 pKa = 4.36GLGVPVASDD128 pKa = 4.2CGLQSPRR135 pKa = 11.84HH136 pKa = 5.51GKK138 pKa = 9.6NIPGISYY145 pKa = 10.62VEE147 pKa = 4.54FEE149 pKa = 4.99FEE151 pKa = 4.06PPPTGSLKK159 pKa = 10.67VIKK162 pKa = 10.17IWDD165 pKa = 3.7DD166 pKa = 3.05QGTGVVYY173 pKa = 10.79DD174 pKa = 3.37GTIDD178 pKa = 3.53VNIYY182 pKa = 10.47DD183 pKa = 4.09SEE185 pKa = 4.43GTLVDD190 pKa = 3.65TLILSDD196 pKa = 3.71ANNWEE201 pKa = 4.41DD202 pKa = 4.1DD203 pKa = 3.98LGYY206 pKa = 10.65LLPGVYY212 pKa = 9.19TFAEE216 pKa = 4.45VTPDD220 pKa = 2.86GWTPSYY226 pKa = 11.35DD227 pKa = 3.4PANRR231 pKa = 11.84EE232 pKa = 4.14LVVEE236 pKa = 4.76AGDD239 pKa = 4.3DD240 pKa = 3.73PAAGAIGTITNTIDD254 pKa = 3.23LGSLKK259 pKa = 10.45VIKK262 pKa = 10.25IWDD265 pKa = 3.7DD266 pKa = 3.05QGTGVVYY273 pKa = 10.79DD274 pKa = 3.37GTIDD278 pKa = 3.53VNIYY282 pKa = 10.47DD283 pKa = 4.09SEE285 pKa = 4.43GTLVDD290 pKa = 3.76TLILSDD296 pKa = 3.69TNNWEE301 pKa = 4.15DD302 pKa = 3.76DD303 pKa = 3.78LGYY306 pKa = 10.65LLPGVYY312 pKa = 9.19TFAEE316 pKa = 4.45VTPDD320 pKa = 2.47GWTPATTRR328 pKa = 11.84QTVSS332 pKa = 2.66
MM1 pKa = 7.28NMAKK5 pKa = 9.96PVSKK9 pKa = 9.91YY10 pKa = 7.29WRR12 pKa = 11.84VGLVLLVIGLLAIGTALGSVPPCEE36 pKa = 4.74GPLVDD41 pKa = 3.48PVEE44 pKa = 4.36YY45 pKa = 10.62NGNPEE50 pKa = 4.31EE51 pKa = 4.35IGTRR55 pKa = 11.84INDD58 pKa = 3.94PGSGYY63 pKa = 9.99IDD65 pKa = 3.52VEE67 pKa = 4.57FDD69 pKa = 3.67GVWYY73 pKa = 10.2KK74 pKa = 10.79VFYY77 pKa = 10.24EE78 pKa = 4.55VYY80 pKa = 10.58DD81 pKa = 3.59GGKK84 pKa = 7.18TLKK87 pKa = 10.04FWEE90 pKa = 4.46EE91 pKa = 3.45NGFPVVSKK99 pKa = 9.12VTVKK103 pKa = 10.77GGPNANIYY111 pKa = 10.26YY112 pKa = 9.79YY113 pKa = 10.94AADD116 pKa = 3.99PPEE119 pKa = 4.36GLGVPVASDD128 pKa = 4.2CGLQSPRR135 pKa = 11.84HH136 pKa = 5.51GKK138 pKa = 9.6NIPGISYY145 pKa = 10.62VEE147 pKa = 4.54FEE149 pKa = 4.99FEE151 pKa = 4.06PPPTGSLKK159 pKa = 10.67VIKK162 pKa = 10.17IWDD165 pKa = 3.7DD166 pKa = 3.05QGTGVVYY173 pKa = 10.79DD174 pKa = 3.37GTIDD178 pKa = 3.53VNIYY182 pKa = 10.47DD183 pKa = 4.09SEE185 pKa = 4.43GTLVDD190 pKa = 3.65TLILSDD196 pKa = 3.71ANNWEE201 pKa = 4.41DD202 pKa = 4.1DD203 pKa = 3.98LGYY206 pKa = 10.65LLPGVYY212 pKa = 9.19TFAEE216 pKa = 4.45VTPDD220 pKa = 2.86GWTPSYY226 pKa = 11.35DD227 pKa = 3.4PANRR231 pKa = 11.84EE232 pKa = 4.14LVVEE236 pKa = 4.76AGDD239 pKa = 4.3DD240 pKa = 3.73PAAGAIGTITNTIDD254 pKa = 3.23LGSLKK259 pKa = 10.45VIKK262 pKa = 10.25IWDD265 pKa = 3.7DD266 pKa = 3.05QGTGVVYY273 pKa = 10.79DD274 pKa = 3.37GTIDD278 pKa = 3.53VNIYY282 pKa = 10.47DD283 pKa = 4.09SEE285 pKa = 4.43GTLVDD290 pKa = 3.76TLILSDD296 pKa = 3.69TNNWEE301 pKa = 4.15DD302 pKa = 3.76DD303 pKa = 3.78LGYY306 pKa = 10.65LLPGVYY312 pKa = 9.19TFAEE316 pKa = 4.45VTPDD320 pKa = 2.47GWTPATTRR328 pKa = 11.84QTVSS332 pKa = 2.66
Molecular weight: 35.9 kDa
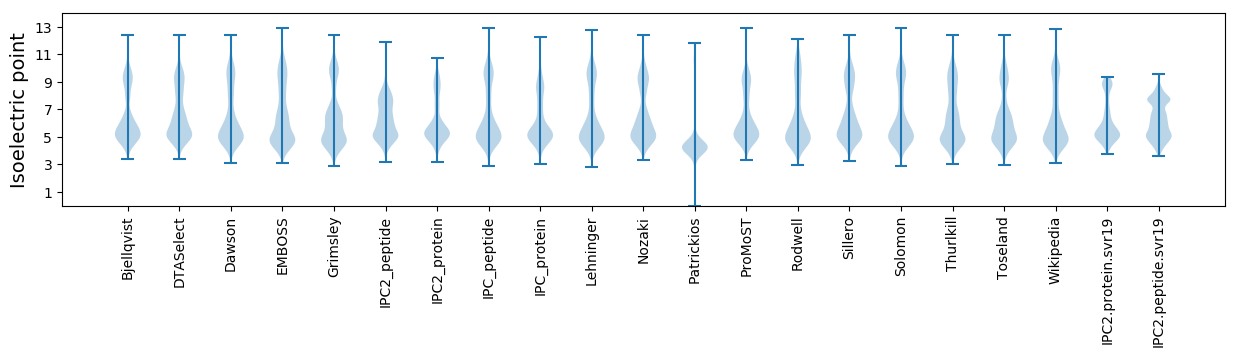
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0GHY1|C0GHY1_9FIRM UspA domain protein OS=Dethiobacter alkaliphilus AHT 1 OX=555088 GN=DealDRAFT_2090 PE=3 SV=1
MM1 pKa = 7.59APKK4 pKa = 9.94KK5 pKa = 10.2IKK7 pKa = 10.47GISGSRR13 pKa = 11.84RR14 pKa = 11.84ATYY17 pKa = 10.77APTTDD22 pKa = 4.81IIPSPPQTCDD32 pKa = 2.23GFMYY36 pKa = 9.99VVRR39 pKa = 11.84PRR41 pKa = 11.84DD42 pKa = 3.47TLYY45 pKa = 11.12SIARR49 pKa = 11.84KK50 pKa = 9.23FNCSVMQLLQANPQISSRR68 pKa = 11.84SGTLFIRR75 pKa = 11.84QRR77 pKa = 11.84ICIPDD82 pKa = 3.62VVDD85 pKa = 3.47VLPAHH90 pKa = 6.99KK91 pKa = 10.03LLPAGPKK98 pKa = 9.71VLFVEE103 pKa = 5.33LLDD106 pKa = 4.64SMGNPLRR113 pKa = 11.84VMNGFVSLAPRR124 pKa = 11.84TFIRR128 pKa = 11.84VVFSEE133 pKa = 4.6PVNQVYY139 pKa = 8.7FFFVPSRR146 pKa = 11.84RR147 pKa = 11.84KK148 pKa = 9.05IFRR151 pKa = 11.84PSFLIGEE158 pKa = 4.44RR159 pKa = 11.84TLVPPSRR166 pKa = 11.84SVRR169 pKa = 11.84FVWDD173 pKa = 3.32VPRR176 pKa = 11.84GIRR179 pKa = 11.84GSLFIVGCTEE189 pKa = 4.94RR190 pKa = 11.84ICGPPEE196 pKa = 3.69EE197 pKa = 4.39LLVRR201 pKa = 11.84RR202 pKa = 11.84PP203 pKa = 3.32
MM1 pKa = 7.59APKK4 pKa = 9.94KK5 pKa = 10.2IKK7 pKa = 10.47GISGSRR13 pKa = 11.84RR14 pKa = 11.84ATYY17 pKa = 10.77APTTDD22 pKa = 4.81IIPSPPQTCDD32 pKa = 2.23GFMYY36 pKa = 9.99VVRR39 pKa = 11.84PRR41 pKa = 11.84DD42 pKa = 3.47TLYY45 pKa = 11.12SIARR49 pKa = 11.84KK50 pKa = 9.23FNCSVMQLLQANPQISSRR68 pKa = 11.84SGTLFIRR75 pKa = 11.84QRR77 pKa = 11.84ICIPDD82 pKa = 3.62VVDD85 pKa = 3.47VLPAHH90 pKa = 6.99KK91 pKa = 10.03LLPAGPKK98 pKa = 9.71VLFVEE103 pKa = 5.33LLDD106 pKa = 4.64SMGNPLRR113 pKa = 11.84VMNGFVSLAPRR124 pKa = 11.84TFIRR128 pKa = 11.84VVFSEE133 pKa = 4.6PVNQVYY139 pKa = 8.7FFFVPSRR146 pKa = 11.84RR147 pKa = 11.84KK148 pKa = 9.05IFRR151 pKa = 11.84PSFLIGEE158 pKa = 4.44RR159 pKa = 11.84TLVPPSRR166 pKa = 11.84SVRR169 pKa = 11.84FVWDD173 pKa = 3.32VPRR176 pKa = 11.84GIRR179 pKa = 11.84GSLFIVGCTEE189 pKa = 4.94RR190 pKa = 11.84ICGPPEE196 pKa = 3.69EE197 pKa = 4.39LLVRR201 pKa = 11.84RR202 pKa = 11.84PP203 pKa = 3.32
Molecular weight: 22.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
920342 |
30 |
1924 |
291.1 |
32.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.726 ± 0.054 | 1.069 ± 0.022 |
5.198 ± 0.038 | 7.376 ± 0.059 |
4.068 ± 0.033 | 7.597 ± 0.045 |
1.958 ± 0.018 | 6.497 ± 0.038 |
5.258 ± 0.043 | 10.43 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.023 | 3.69 ± 0.033 |
4.147 ± 0.029 | 3.74 ± 0.027 |
5.241 ± 0.037 | 5.319 ± 0.029 |
5.167 ± 0.029 | 7.741 ± 0.038 |
0.932 ± 0.017 | 3.103 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
