
Labedaea rhizosphaerae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Labedaea
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
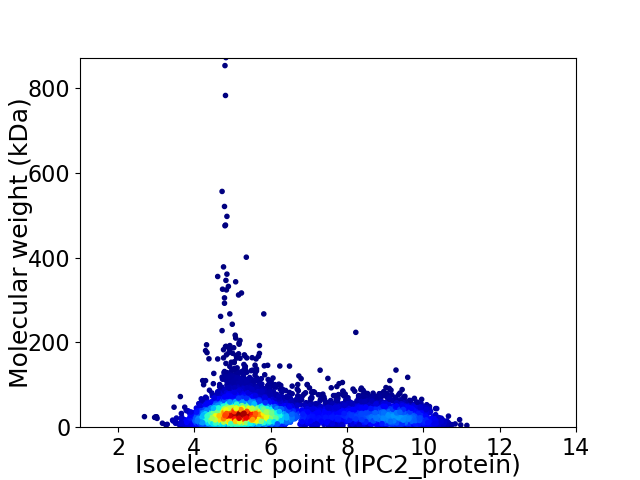
Virtual 2D-PAGE plot for 7717 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6SGB8|A0A4R6SGB8_9PSEU Pimeloyl-ACP methyl ester carboxylesterase OS=Labedaea rhizosphaerae OX=598644 GN=EV186_102448 PE=4 SV=1
MM1 pKa = 7.34SLDD4 pKa = 3.61PTTLQSFVTNLLTDD18 pKa = 4.03DD19 pKa = 4.03AARR22 pKa = 11.84SAFAADD28 pKa = 3.6PTAVLQGAGLGDD40 pKa = 3.58ITPQDD45 pKa = 3.87VQQVIPLVTDD55 pKa = 3.12QLPGLDD61 pKa = 4.17GGLPTLPALPTEE73 pKa = 4.33QVNDD77 pKa = 3.82AVAQLQAIADD87 pKa = 3.86AAQANSGLPLPHH99 pKa = 7.35VGFASTSDD107 pKa = 3.28QGNLAGAVTVTGDD120 pKa = 3.14GVTGGGAASAGVDD133 pKa = 3.55GVGVTGGVDD142 pKa = 3.21SAYY145 pKa = 9.41GTAHH149 pKa = 6.54GSGVVGLDD157 pKa = 3.06GGEE160 pKa = 4.42GNLTAASHH168 pKa = 6.06QLHH171 pKa = 5.94TWSASDD177 pKa = 3.96ASVGLDD183 pKa = 3.14GAAAATSTHH192 pKa = 6.1VYY194 pKa = 10.86DD195 pKa = 4.05NAVSGAATVTPDD207 pKa = 3.33GNFTAVGGAHH217 pKa = 6.84SDD219 pKa = 3.27LGDD222 pKa = 3.32VHH224 pKa = 6.6GAVAGGLDD232 pKa = 3.48GVYY235 pKa = 10.92GNGGVHH241 pKa = 6.62TDD243 pKa = 4.09DD244 pKa = 5.27LATNTDD250 pKa = 3.93FHH252 pKa = 6.34ATQDD256 pKa = 3.43GFGVVTNGYY265 pKa = 9.73GSQLGSFQSAAAGSTDD281 pKa = 3.47RR282 pKa = 11.84VATHH286 pKa = 7.0NEE288 pKa = 3.77FQTANAQGSNDD299 pKa = 3.3FSVGKK304 pKa = 10.3DD305 pKa = 3.37GLATSSTLQTDD316 pKa = 3.9QVSAASDD323 pKa = 3.46ATVGTDD329 pKa = 3.34GTFTSTTAADD339 pKa = 3.65SQFGGFSATASGSTQGFAGATEE361 pKa = 3.83FHH363 pKa = 6.54GAGVNGGGGVAAGLDD378 pKa = 3.91GVTGKK383 pKa = 10.61AGLDD387 pKa = 3.47SQFGHH392 pKa = 6.48VDD394 pKa = 3.04VAGAVSTSGGAGALAVGNDD413 pKa = 3.5MLGVEE418 pKa = 4.68GDD420 pKa = 3.8VAATTDD426 pKa = 3.84SVSLGGDD433 pKa = 3.54LHH435 pKa = 6.49TPAGDD440 pKa = 4.04LPLTPVNIPVGLPGVDD456 pKa = 3.45GLPGVGGLTGALPGVGALPGVDD478 pKa = 3.89GLTGALPGVDD488 pKa = 4.08GLTGALPVDD497 pKa = 4.26GLEE500 pKa = 4.54HH501 pKa = 6.95GLPDD505 pKa = 3.85VDD507 pKa = 4.18GLTGAVPGVDD517 pKa = 3.77GLTGALPLDD526 pKa = 4.04GLTNGGLPGVDD537 pKa = 3.71GLTGALPVDD546 pKa = 4.26ALSGALPLADD556 pKa = 3.36QFGGLGDD563 pKa = 3.9PAHH566 pKa = 6.86ILDD569 pKa = 4.67SIEE572 pKa = 3.81PARR575 pKa = 11.84GGEE578 pKa = 4.02AAVADD583 pKa = 4.15YY584 pKa = 10.94VSNGGQVLTDD594 pKa = 3.7GLATGTNTLGQYY606 pKa = 8.44LTGTPAAPVADD617 pKa = 3.84VVEE620 pKa = 4.66TGSNTVSGIVSQGADD635 pKa = 3.29VASHH639 pKa = 6.13VPALPAVSDD648 pKa = 4.33LPSHH652 pKa = 6.65LPVGDD657 pKa = 4.62LPAASDD663 pKa = 4.22LPNLSDD669 pKa = 4.71LPSHH673 pKa = 6.89LPVDD677 pKa = 4.57LPNLPVANPLPEE689 pKa = 4.3VTQHH693 pKa = 4.93VTDD696 pKa = 4.27LTSHH700 pKa = 5.26VTDD703 pKa = 3.39VTSTVGNVTDD713 pKa = 3.96VVTHH717 pKa = 6.1NPVTDD722 pKa = 4.01VVSHH726 pKa = 6.35SPLGNVTGGDD736 pKa = 3.4HH737 pKa = 6.86GLLGGDD743 pKa = 3.62GLLPDD748 pKa = 4.54LHH750 pKa = 7.48LGHH753 pKa = 7.41
MM1 pKa = 7.34SLDD4 pKa = 3.61PTTLQSFVTNLLTDD18 pKa = 4.03DD19 pKa = 4.03AARR22 pKa = 11.84SAFAADD28 pKa = 3.6PTAVLQGAGLGDD40 pKa = 3.58ITPQDD45 pKa = 3.87VQQVIPLVTDD55 pKa = 3.12QLPGLDD61 pKa = 4.17GGLPTLPALPTEE73 pKa = 4.33QVNDD77 pKa = 3.82AVAQLQAIADD87 pKa = 3.86AAQANSGLPLPHH99 pKa = 7.35VGFASTSDD107 pKa = 3.28QGNLAGAVTVTGDD120 pKa = 3.14GVTGGGAASAGVDD133 pKa = 3.55GVGVTGGVDD142 pKa = 3.21SAYY145 pKa = 9.41GTAHH149 pKa = 6.54GSGVVGLDD157 pKa = 3.06GGEE160 pKa = 4.42GNLTAASHH168 pKa = 6.06QLHH171 pKa = 5.94TWSASDD177 pKa = 3.96ASVGLDD183 pKa = 3.14GAAAATSTHH192 pKa = 6.1VYY194 pKa = 10.86DD195 pKa = 4.05NAVSGAATVTPDD207 pKa = 3.33GNFTAVGGAHH217 pKa = 6.84SDD219 pKa = 3.27LGDD222 pKa = 3.32VHH224 pKa = 6.6GAVAGGLDD232 pKa = 3.48GVYY235 pKa = 10.92GNGGVHH241 pKa = 6.62TDD243 pKa = 4.09DD244 pKa = 5.27LATNTDD250 pKa = 3.93FHH252 pKa = 6.34ATQDD256 pKa = 3.43GFGVVTNGYY265 pKa = 9.73GSQLGSFQSAAAGSTDD281 pKa = 3.47RR282 pKa = 11.84VATHH286 pKa = 7.0NEE288 pKa = 3.77FQTANAQGSNDD299 pKa = 3.3FSVGKK304 pKa = 10.3DD305 pKa = 3.37GLATSSTLQTDD316 pKa = 3.9QVSAASDD323 pKa = 3.46ATVGTDD329 pKa = 3.34GTFTSTTAADD339 pKa = 3.65SQFGGFSATASGSTQGFAGATEE361 pKa = 3.83FHH363 pKa = 6.54GAGVNGGGGVAAGLDD378 pKa = 3.91GVTGKK383 pKa = 10.61AGLDD387 pKa = 3.47SQFGHH392 pKa = 6.48VDD394 pKa = 3.04VAGAVSTSGGAGALAVGNDD413 pKa = 3.5MLGVEE418 pKa = 4.68GDD420 pKa = 3.8VAATTDD426 pKa = 3.84SVSLGGDD433 pKa = 3.54LHH435 pKa = 6.49TPAGDD440 pKa = 4.04LPLTPVNIPVGLPGVDD456 pKa = 3.45GLPGVGGLTGALPGVGALPGVDD478 pKa = 3.89GLTGALPGVDD488 pKa = 4.08GLTGALPVDD497 pKa = 4.26GLEE500 pKa = 4.54HH501 pKa = 6.95GLPDD505 pKa = 3.85VDD507 pKa = 4.18GLTGAVPGVDD517 pKa = 3.77GLTGALPLDD526 pKa = 4.04GLTNGGLPGVDD537 pKa = 3.71GLTGALPVDD546 pKa = 4.26ALSGALPLADD556 pKa = 3.36QFGGLGDD563 pKa = 3.9PAHH566 pKa = 6.86ILDD569 pKa = 4.67SIEE572 pKa = 3.81PARR575 pKa = 11.84GGEE578 pKa = 4.02AAVADD583 pKa = 4.15YY584 pKa = 10.94VSNGGQVLTDD594 pKa = 3.7GLATGTNTLGQYY606 pKa = 8.44LTGTPAAPVADD617 pKa = 3.84VVEE620 pKa = 4.66TGSNTVSGIVSQGADD635 pKa = 3.29VASHH639 pKa = 6.13VPALPAVSDD648 pKa = 4.33LPSHH652 pKa = 6.65LPVGDD657 pKa = 4.62LPAASDD663 pKa = 4.22LPNLSDD669 pKa = 4.71LPSHH673 pKa = 6.89LPVDD677 pKa = 4.57LPNLPVANPLPEE689 pKa = 4.3VTQHH693 pKa = 4.93VTDD696 pKa = 4.27LTSHH700 pKa = 5.26VTDD703 pKa = 3.39VTSTVGNVTDD713 pKa = 3.96VVTHH717 pKa = 6.1NPVTDD722 pKa = 4.01VVSHH726 pKa = 6.35SPLGNVTGGDD736 pKa = 3.4HH737 pKa = 6.86GLLGGDD743 pKa = 3.62GLLPDD748 pKa = 4.54LHH750 pKa = 7.48LGHH753 pKa = 7.41
Molecular weight: 72.25 kDa
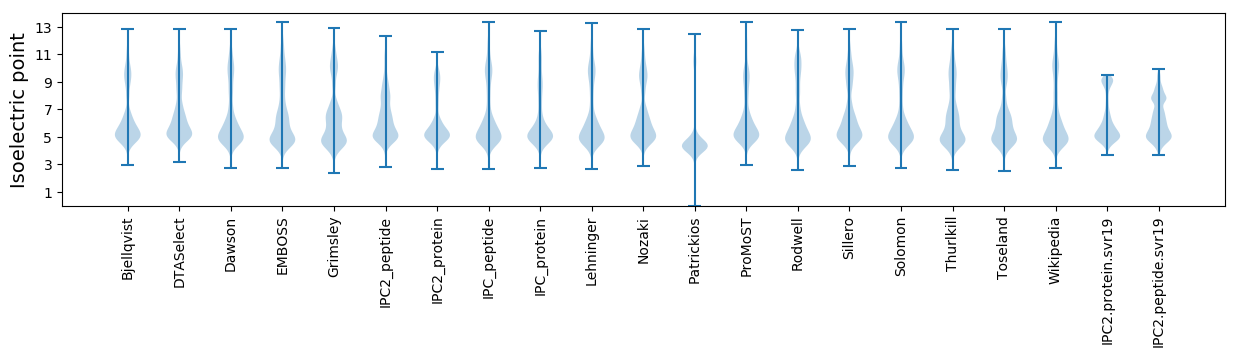
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6SRF3|A0A4R6SRF3_9PSEU Bifunctional purine biosynthesis protein PurH OS=Labedaea rhizosphaerae OX=598644 GN=purH PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.46LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.97LKK32 pKa = 10.51KK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.46LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.97LKK32 pKa = 10.51KK33 pKa = 9.87
Molecular weight: 4.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2529000 |
29 |
8347 |
327.7 |
35.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.492 ± 0.049 | 0.791 ± 0.008 |
6.333 ± 0.023 | 5.268 ± 0.027 |
2.795 ± 0.015 | 9.104 ± 0.032 |
2.3 ± 0.013 | 3.397 ± 0.02 |
2.176 ± 0.023 | 10.34 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.798 ± 0.012 | 1.926 ± 0.016 |
5.874 ± 0.025 | 2.977 ± 0.018 |
7.516 ± 0.036 | 5.016 ± 0.021 |
6.262 ± 0.028 | 9.116 ± 0.029 |
1.545 ± 0.011 | 1.976 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
