
Bacillus phage W.Ph.
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Herelleviridae; Bastillevirinae; Wphvirus; Bacillus virus WPh
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
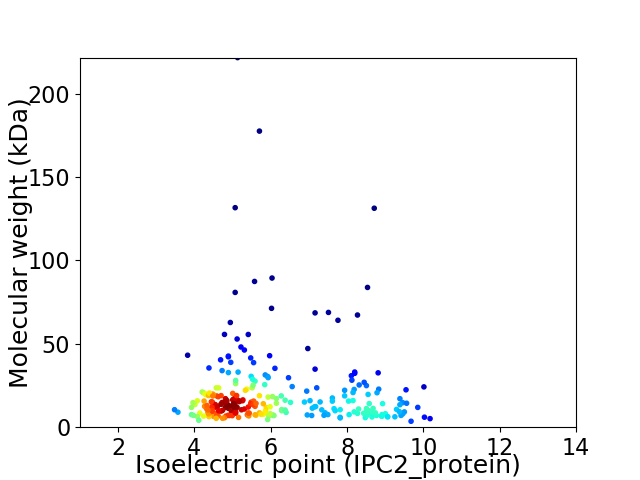
Virtual 2D-PAGE plot for 274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9B1S8|G9B1S8_9CAUD Gp177 OS=Bacillus phage W.Ph. OX=764595 PE=4 SV=1
MM1 pKa = 7.85RR2 pKa = 11.84MKK4 pKa = 10.57KK5 pKa = 9.86VDD7 pKa = 3.68KK8 pKa = 9.53LTAAIEE14 pKa = 4.2MVKK17 pKa = 10.5GLEE20 pKa = 4.32VKK22 pKa = 10.78CGGLEE27 pKa = 4.22VNWLEE32 pKa = 4.78STDD35 pKa = 3.36EE36 pKa = 3.92WVINYY41 pKa = 9.97GDD43 pKa = 4.32VIIMDD48 pKa = 5.09DD49 pKa = 3.61LTEE52 pKa = 4.07EE53 pKa = 3.95QAYY56 pKa = 8.99EE57 pKa = 4.01CHH59 pKa = 6.1GTLTQYY65 pKa = 9.98EE66 pKa = 4.47VQGVQVEE73 pKa = 4.38LKK75 pKa = 10.04EE76 pKa = 4.09YY77 pKa = 9.91NYY79 pKa = 10.9DD80 pKa = 3.4YY81 pKa = 11.89AMEE84 pKa = 4.41NNKK87 pKa = 10.11GEE89 pKa = 4.39SFTEE93 pKa = 3.7IALEE97 pKa = 4.5RR98 pKa = 11.84MVIDD102 pKa = 3.64DD103 pKa = 3.99WGSDD107 pKa = 3.46FEE109 pKa = 4.62TVAFEE114 pKa = 4.29VADD117 pKa = 4.0GVTPFANYY125 pKa = 9.15EE126 pKa = 3.7IWEE129 pKa = 4.4VASNNYY135 pKa = 9.51EE136 pKa = 4.22SIEE139 pKa = 4.0EE140 pKa = 4.21NIEE143 pKa = 3.63EE144 pKa = 4.18GLIVIDD150 pKa = 5.19GEE152 pKa = 4.39LNKK155 pKa = 10.7NFMASIAYY163 pKa = 7.44TLTRR167 pKa = 11.84SIEE170 pKa = 4.04QNKK173 pKa = 10.1EE174 pKa = 3.79EE175 pKa = 4.32ILYY178 pKa = 10.6NYY180 pKa = 8.98IAYY183 pKa = 9.59RR184 pKa = 11.84VNEE187 pKa = 4.07MLVKK191 pKa = 10.53KK192 pKa = 10.37RR193 pKa = 11.84VTDD196 pKa = 3.71EE197 pKa = 4.46NIIEE201 pKa = 4.11YY202 pKa = 10.63LEE204 pKa = 4.12EE205 pKa = 4.52EE206 pKa = 4.22IEE208 pKa = 4.27NYY210 pKa = 10.51ADD212 pKa = 3.53EE213 pKa = 5.45AEE215 pKa = 4.08NSYY218 pKa = 11.68NEE220 pKa = 4.11FNYY223 pKa = 10.68LDD225 pKa = 4.85DD226 pKa = 4.61EE227 pKa = 4.49LFEE230 pKa = 4.03QLEE233 pKa = 4.34EE234 pKa = 5.06FIDD237 pKa = 4.05SYY239 pKa = 11.41IAGLSMGEE247 pKa = 4.02SLGNQDD253 pKa = 3.85YY254 pKa = 11.45EE255 pKa = 4.58NIGDD259 pKa = 3.78VTVTEE264 pKa = 4.23GAMFIKK270 pKa = 10.26KK271 pKa = 10.43DD272 pKa = 3.3EE273 pKa = 4.62DD274 pKa = 3.78TDD276 pKa = 3.24KK277 pKa = 10.92SYY279 pKa = 11.66YY280 pKa = 8.52VVRR283 pKa = 11.84VCGNEE288 pKa = 3.35FDD290 pKa = 5.12GFYY293 pKa = 10.27LDD295 pKa = 5.47SMYY298 pKa = 10.73IDD300 pKa = 5.85LNDD303 pKa = 3.99GWIDD307 pKa = 3.25WYY309 pKa = 10.94AINDD313 pKa = 3.63YY314 pKa = 11.38AGIDD318 pKa = 3.78MDD320 pKa = 4.89ADD322 pKa = 3.67DD323 pKa = 4.56YY324 pKa = 11.87EE325 pKa = 4.49RR326 pKa = 11.84VNALISYY333 pKa = 10.25DD334 pKa = 4.03GIDD337 pKa = 3.78NFNPSTTEE345 pKa = 3.67CKK347 pKa = 9.39TAIDD351 pKa = 3.67LCRR354 pKa = 11.84ALRR357 pKa = 11.84EE358 pKa = 4.1AGVNVVWSAEE368 pKa = 3.69HH369 pKa = 7.51SEE371 pKa = 4.07IQFF374 pKa = 3.45
MM1 pKa = 7.85RR2 pKa = 11.84MKK4 pKa = 10.57KK5 pKa = 9.86VDD7 pKa = 3.68KK8 pKa = 9.53LTAAIEE14 pKa = 4.2MVKK17 pKa = 10.5GLEE20 pKa = 4.32VKK22 pKa = 10.78CGGLEE27 pKa = 4.22VNWLEE32 pKa = 4.78STDD35 pKa = 3.36EE36 pKa = 3.92WVINYY41 pKa = 9.97GDD43 pKa = 4.32VIIMDD48 pKa = 5.09DD49 pKa = 3.61LTEE52 pKa = 4.07EE53 pKa = 3.95QAYY56 pKa = 8.99EE57 pKa = 4.01CHH59 pKa = 6.1GTLTQYY65 pKa = 9.98EE66 pKa = 4.47VQGVQVEE73 pKa = 4.38LKK75 pKa = 10.04EE76 pKa = 4.09YY77 pKa = 9.91NYY79 pKa = 10.9DD80 pKa = 3.4YY81 pKa = 11.89AMEE84 pKa = 4.41NNKK87 pKa = 10.11GEE89 pKa = 4.39SFTEE93 pKa = 3.7IALEE97 pKa = 4.5RR98 pKa = 11.84MVIDD102 pKa = 3.64DD103 pKa = 3.99WGSDD107 pKa = 3.46FEE109 pKa = 4.62TVAFEE114 pKa = 4.29VADD117 pKa = 4.0GVTPFANYY125 pKa = 9.15EE126 pKa = 3.7IWEE129 pKa = 4.4VASNNYY135 pKa = 9.51EE136 pKa = 4.22SIEE139 pKa = 4.0EE140 pKa = 4.21NIEE143 pKa = 3.63EE144 pKa = 4.18GLIVIDD150 pKa = 5.19GEE152 pKa = 4.39LNKK155 pKa = 10.7NFMASIAYY163 pKa = 7.44TLTRR167 pKa = 11.84SIEE170 pKa = 4.04QNKK173 pKa = 10.1EE174 pKa = 3.79EE175 pKa = 4.32ILYY178 pKa = 10.6NYY180 pKa = 8.98IAYY183 pKa = 9.59RR184 pKa = 11.84VNEE187 pKa = 4.07MLVKK191 pKa = 10.53KK192 pKa = 10.37RR193 pKa = 11.84VTDD196 pKa = 3.71EE197 pKa = 4.46NIIEE201 pKa = 4.11YY202 pKa = 10.63LEE204 pKa = 4.12EE205 pKa = 4.52EE206 pKa = 4.22IEE208 pKa = 4.27NYY210 pKa = 10.51ADD212 pKa = 3.53EE213 pKa = 5.45AEE215 pKa = 4.08NSYY218 pKa = 11.68NEE220 pKa = 4.11FNYY223 pKa = 10.68LDD225 pKa = 4.85DD226 pKa = 4.61EE227 pKa = 4.49LFEE230 pKa = 4.03QLEE233 pKa = 4.34EE234 pKa = 5.06FIDD237 pKa = 4.05SYY239 pKa = 11.41IAGLSMGEE247 pKa = 4.02SLGNQDD253 pKa = 3.85YY254 pKa = 11.45EE255 pKa = 4.58NIGDD259 pKa = 3.78VTVTEE264 pKa = 4.23GAMFIKK270 pKa = 10.26KK271 pKa = 10.43DD272 pKa = 3.3EE273 pKa = 4.62DD274 pKa = 3.78TDD276 pKa = 3.24KK277 pKa = 10.92SYY279 pKa = 11.66YY280 pKa = 8.52VVRR283 pKa = 11.84VCGNEE288 pKa = 3.35FDD290 pKa = 5.12GFYY293 pKa = 10.27LDD295 pKa = 5.47SMYY298 pKa = 10.73IDD300 pKa = 5.85LNDD303 pKa = 3.99GWIDD307 pKa = 3.25WYY309 pKa = 10.94AINDD313 pKa = 3.63YY314 pKa = 11.38AGIDD318 pKa = 3.78MDD320 pKa = 4.89ADD322 pKa = 3.67DD323 pKa = 4.56YY324 pKa = 11.87EE325 pKa = 4.49RR326 pKa = 11.84VNALISYY333 pKa = 10.25DD334 pKa = 4.03GIDD337 pKa = 3.78NFNPSTTEE345 pKa = 3.67CKK347 pKa = 9.39TAIDD351 pKa = 3.67LCRR354 pKa = 11.84ALRR357 pKa = 11.84EE358 pKa = 4.1AGVNVVWSAEE368 pKa = 3.69HH369 pKa = 7.51SEE371 pKa = 4.07IQFF374 pKa = 3.45
Molecular weight: 43.16 kDa
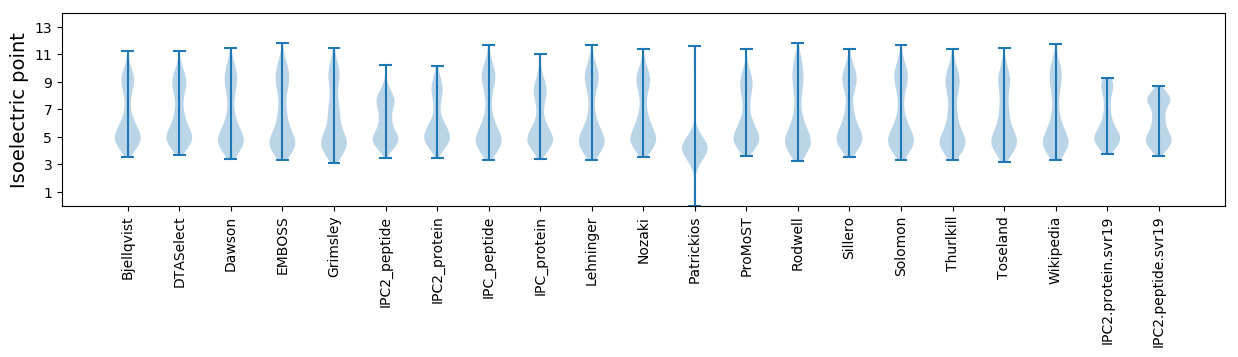
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9B1C1|G9B1C1_9CAUD Gp20 OS=Bacillus phage W.Ph. OX=764595 PE=4 SV=1
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 8.54PRR6 pKa = 11.84KK7 pKa = 8.19PRR9 pKa = 11.84KK10 pKa = 9.35VPKK13 pKa = 9.95LLQGGQEE20 pKa = 3.93TSRR23 pKa = 11.84AINNIGKK30 pKa = 9.61ALQQQALQSTANQLSKK46 pKa = 10.87SLPEE50 pKa = 4.41GYY52 pKa = 9.51EE53 pKa = 3.66LKK55 pKa = 10.39QRR57 pKa = 11.84PKK59 pKa = 10.12YY60 pKa = 10.98LEE62 pKa = 3.91VTEE65 pKa = 4.42KK66 pKa = 10.77RR67 pKa = 11.84LIGMGVIDD75 pKa = 6.02LKK77 pKa = 11.22PLFAKK82 pKa = 10.36SSKK85 pKa = 10.61KK86 pKa = 10.34KK87 pKa = 10.09FSKK90 pKa = 10.45KK91 pKa = 10.26GGWYY95 pKa = 9.93LDD97 pKa = 3.03IPIRR101 pKa = 11.84RR102 pKa = 11.84KK103 pKa = 9.96ARR105 pKa = 11.84GMTRR109 pKa = 11.84RR110 pKa = 11.84MYY112 pKa = 10.55DD113 pKa = 2.9QLRR116 pKa = 11.84AVDD119 pKa = 3.91MGGQQKK125 pKa = 10.4INIISDD131 pKa = 3.46YY132 pKa = 10.78LYY134 pKa = 10.86DD135 pKa = 3.68NRR137 pKa = 11.84GVSDD141 pKa = 4.72APLLNYY147 pKa = 9.51KK148 pKa = 9.89PKK150 pKa = 10.98SNVITKK156 pKa = 9.75IASGKK161 pKa = 9.42GRR163 pKa = 11.84HH164 pKa = 6.64DD165 pKa = 3.29YY166 pKa = 10.67VAFRR170 pKa = 11.84RR171 pKa = 11.84VSNNSSPSSWIINRR185 pKa = 11.84DD186 pKa = 3.12RR187 pKa = 11.84VTVANTSKK195 pKa = 9.8TFVANVNRR203 pKa = 11.84LMKK206 pKa = 10.94YY207 pKa = 9.52NMKK210 pKa = 10.75NMM212 pKa = 3.89
MM1 pKa = 7.27ARR3 pKa = 11.84KK4 pKa = 8.54PRR6 pKa = 11.84KK7 pKa = 8.19PRR9 pKa = 11.84KK10 pKa = 9.35VPKK13 pKa = 9.95LLQGGQEE20 pKa = 3.93TSRR23 pKa = 11.84AINNIGKK30 pKa = 9.61ALQQQALQSTANQLSKK46 pKa = 10.87SLPEE50 pKa = 4.41GYY52 pKa = 9.51EE53 pKa = 3.66LKK55 pKa = 10.39QRR57 pKa = 11.84PKK59 pKa = 10.12YY60 pKa = 10.98LEE62 pKa = 3.91VTEE65 pKa = 4.42KK66 pKa = 10.77RR67 pKa = 11.84LIGMGVIDD75 pKa = 6.02LKK77 pKa = 11.22PLFAKK82 pKa = 10.36SSKK85 pKa = 10.61KK86 pKa = 10.34KK87 pKa = 10.09FSKK90 pKa = 10.45KK91 pKa = 10.26GGWYY95 pKa = 9.93LDD97 pKa = 3.03IPIRR101 pKa = 11.84RR102 pKa = 11.84KK103 pKa = 9.96ARR105 pKa = 11.84GMTRR109 pKa = 11.84RR110 pKa = 11.84MYY112 pKa = 10.55DD113 pKa = 2.9QLRR116 pKa = 11.84AVDD119 pKa = 3.91MGGQQKK125 pKa = 10.4INIISDD131 pKa = 3.46YY132 pKa = 10.78LYY134 pKa = 10.86DD135 pKa = 3.68NRR137 pKa = 11.84GVSDD141 pKa = 4.72APLLNYY147 pKa = 9.51KK148 pKa = 9.89PKK150 pKa = 10.98SNVITKK156 pKa = 9.75IASGKK161 pKa = 9.42GRR163 pKa = 11.84HH164 pKa = 6.64DD165 pKa = 3.29YY166 pKa = 10.67VAFRR170 pKa = 11.84RR171 pKa = 11.84VSNNSSPSSWIINRR185 pKa = 11.84DD186 pKa = 3.12RR187 pKa = 11.84VTVANTSKK195 pKa = 9.8TFVANVNRR203 pKa = 11.84LMKK206 pKa = 10.94YY207 pKa = 9.52NMKK210 pKa = 10.75NMM212 pKa = 3.89
Molecular weight: 24.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
48101 |
32 |
1978 |
175.6 |
20.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.491 ± 0.241 | 0.902 ± 0.078 |
6.559 ± 0.149 | 8.27 ± 0.29 |
3.929 ± 0.087 | 5.969 ± 0.255 |
1.867 ± 0.09 | 6.767 ± 0.134 |
8.224 ± 0.172 | 7.79 ± 0.159 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.114 ± 0.093 | 5.736 ± 0.161 |
2.761 ± 0.129 | 3.056 ± 0.124 |
4.447 ± 0.106 | 5.969 ± 0.129 |
6.33 ± 0.206 | 7.131 ± 0.157 |
1.156 ± 0.066 | 4.532 ± 0.12 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
