
Gyrovirus Tu789
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Gyrovirus homsa2
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
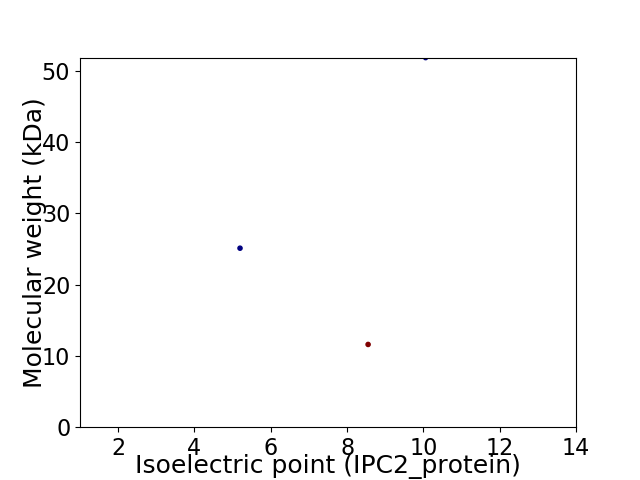
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5UAJ3|U5UAJ3_9VIRU Apoptin OS=Gyrovirus Tu789 OX=1415628 PE=3 SV=1
MM1 pKa = 7.57SSGGVEE7 pKa = 3.92STPPGEE13 pKa = 4.05TRR15 pKa = 11.84PAGGSEE21 pKa = 4.1SPFRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.91GQRR30 pKa = 11.84GPSGAGYY37 pKa = 9.29PGEE40 pKa = 4.26TLTPSPRR47 pKa = 11.84GHH49 pKa = 5.22VRR51 pKa = 11.84KK52 pKa = 9.96HH53 pKa = 5.59IDD55 pKa = 4.23GIVASNKK62 pKa = 9.1FVSVGWDD69 pKa = 3.08SLQRR73 pKa = 11.84DD74 pKa = 4.39PNWTRR79 pKa = 11.84QCYY82 pKa = 9.25NRR84 pKa = 11.84AIASWLRR91 pKa = 11.84NCARR95 pKa = 11.84THH97 pKa = 7.67DD98 pKa = 4.69EE99 pKa = 4.4ICNCGQWRR107 pKa = 11.84RR108 pKa = 11.84HH109 pKa = 4.91WFQEE113 pKa = 4.31CAGLEE118 pKa = 4.22DD119 pKa = 5.15ASSQTEE125 pKa = 4.03DD126 pKa = 3.33PRR128 pKa = 11.84AQEE131 pKa = 3.71DD132 pKa = 3.66LRR134 pKa = 11.84RR135 pKa = 11.84LRR137 pKa = 11.84EE138 pKa = 3.79SGAAAKK144 pKa = 10.27RR145 pKa = 11.84KK146 pKa = 9.47LDD148 pKa = 4.0YY149 pKa = 10.43IDD151 pKa = 3.87RR152 pKa = 11.84CKK154 pKa = 10.58KK155 pKa = 10.06RR156 pKa = 11.84PKK158 pKa = 9.0TVTWFDD164 pKa = 3.24TGEE167 pKa = 4.11EE168 pKa = 4.16PDD170 pKa = 3.62VPEE173 pKa = 4.95GFFTPSEE180 pKa = 4.47DD181 pKa = 3.43EE182 pKa = 4.79DD183 pKa = 6.34GYY185 pKa = 10.88DD186 pKa = 3.43TDD188 pKa = 3.51VDD190 pKa = 3.64EE191 pKa = 5.35DD192 pKa = 4.07AVPGGVNFDD201 pKa = 3.95MRR203 pKa = 11.84VEE205 pKa = 4.26DD206 pKa = 4.09PFLNALRR213 pKa = 11.84GGSTTRR219 pKa = 11.84IQGPTWW225 pKa = 3.22
MM1 pKa = 7.57SSGGVEE7 pKa = 3.92STPPGEE13 pKa = 4.05TRR15 pKa = 11.84PAGGSEE21 pKa = 4.1SPFRR25 pKa = 11.84RR26 pKa = 11.84EE27 pKa = 3.91GQRR30 pKa = 11.84GPSGAGYY37 pKa = 9.29PGEE40 pKa = 4.26TLTPSPRR47 pKa = 11.84GHH49 pKa = 5.22VRR51 pKa = 11.84KK52 pKa = 9.96HH53 pKa = 5.59IDD55 pKa = 4.23GIVASNKK62 pKa = 9.1FVSVGWDD69 pKa = 3.08SLQRR73 pKa = 11.84DD74 pKa = 4.39PNWTRR79 pKa = 11.84QCYY82 pKa = 9.25NRR84 pKa = 11.84AIASWLRR91 pKa = 11.84NCARR95 pKa = 11.84THH97 pKa = 7.67DD98 pKa = 4.69EE99 pKa = 4.4ICNCGQWRR107 pKa = 11.84RR108 pKa = 11.84HH109 pKa = 4.91WFQEE113 pKa = 4.31CAGLEE118 pKa = 4.22DD119 pKa = 5.15ASSQTEE125 pKa = 4.03DD126 pKa = 3.33PRR128 pKa = 11.84AQEE131 pKa = 3.71DD132 pKa = 3.66LRR134 pKa = 11.84RR135 pKa = 11.84LRR137 pKa = 11.84EE138 pKa = 3.79SGAAAKK144 pKa = 10.27RR145 pKa = 11.84KK146 pKa = 9.47LDD148 pKa = 4.0YY149 pKa = 10.43IDD151 pKa = 3.87RR152 pKa = 11.84CKK154 pKa = 10.58KK155 pKa = 10.06RR156 pKa = 11.84PKK158 pKa = 9.0TVTWFDD164 pKa = 3.24TGEE167 pKa = 4.11EE168 pKa = 4.16PDD170 pKa = 3.62VPEE173 pKa = 4.95GFFTPSEE180 pKa = 4.47DD181 pKa = 3.43EE182 pKa = 4.79DD183 pKa = 6.34GYY185 pKa = 10.88DD186 pKa = 3.43TDD188 pKa = 3.51VDD190 pKa = 3.64EE191 pKa = 5.35DD192 pKa = 4.07AVPGGVNFDD201 pKa = 3.95MRR203 pKa = 11.84VEE205 pKa = 4.26DD206 pKa = 4.09PFLNALRR213 pKa = 11.84GGSTTRR219 pKa = 11.84IQGPTWW225 pKa = 3.22
Molecular weight: 25.12 kDa
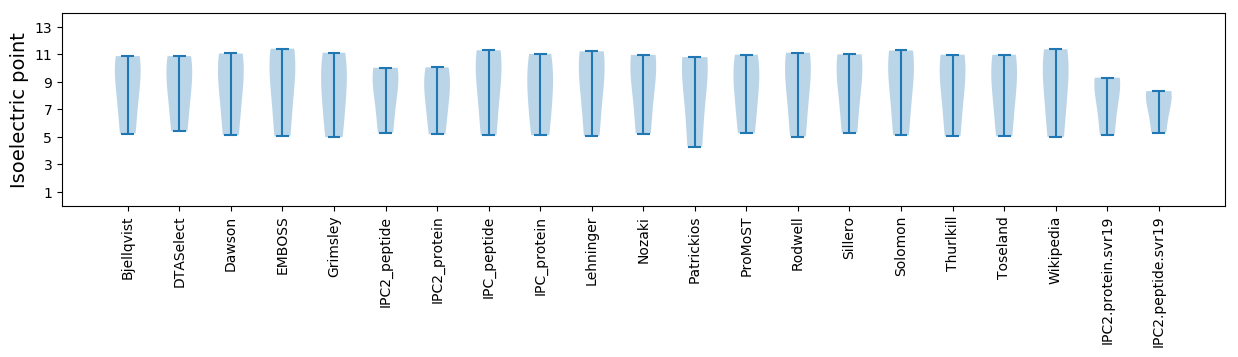
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5UA55|U5UA55_9VIRU Dual specificity protein phosphatase VP2 OS=Gyrovirus Tu789 OX=1415628 PE=3 SV=1
MM1 pKa = 6.6VRR3 pKa = 11.84YY4 pKa = 9.35RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84GVFYY16 pKa = 11.07SLGRR20 pKa = 11.84RR21 pKa = 11.84GWIRR25 pKa = 11.84HH26 pKa = 3.82RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84TWWGKK38 pKa = 8.44FRR40 pKa = 11.84HH41 pKa = 5.05ARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84SFSQRR50 pKa = 11.84VKK52 pKa = 10.66RR53 pKa = 11.84RR54 pKa = 11.84FHH56 pKa = 6.18NPHH59 pKa = 6.89PGTYY63 pKa = 9.85LVRR66 pKa = 11.84LPRR69 pKa = 11.84PQTTLTIFFQGIVYY83 pKa = 9.57FPTAKK88 pKa = 9.68MYY90 pKa = 10.85ASRR93 pKa = 11.84NQLADD98 pKa = 3.63LNKK101 pKa = 9.23ITTCRR106 pKa = 11.84VGLMNINFRR115 pKa = 11.84EE116 pKa = 4.14FLLAALPLDD125 pKa = 4.57AYY127 pKa = 10.77SKK129 pKa = 10.87LGGPIVGPQYY139 pKa = 11.15VSGGGSSAGTHH150 pKa = 7.04DD151 pKa = 3.34EE152 pKa = 4.4TMWPFTARR160 pKa = 11.84PVTNKK165 pKa = 9.83QPGASPAEE173 pKa = 3.89WWRR176 pKa = 11.84WALLLMFPRR185 pKa = 11.84EE186 pKa = 3.78PNMFFRR192 pKa = 11.84QPQQPTLEE200 pKa = 4.06QMGEE204 pKa = 4.01IFGGWQLYY212 pKa = 8.37RR213 pKa = 11.84HH214 pKa = 6.77IKK216 pKa = 8.46TKK218 pKa = 9.5VAIMAAQDD226 pKa = 3.32GGAFSPVASLVSQNNYY242 pKa = 6.08FWRR245 pKa = 11.84RR246 pKa = 11.84QEE248 pKa = 3.83RR249 pKa = 11.84GTPKK253 pKa = 10.0TGEE256 pKa = 3.9PPMTGMIRR264 pKa = 11.84RR265 pKa = 11.84GTGSLEE271 pKa = 3.89SSKK274 pKa = 10.8PADD277 pKa = 3.74EE278 pKa = 4.79YY279 pKa = 11.38YY280 pKa = 10.3IPANPPNPPDD290 pKa = 3.38WPGEE294 pKa = 4.03SSAVIDD300 pKa = 3.91TPYY303 pKa = 10.56KK304 pKa = 9.35IAAVRR309 pKa = 11.84SKK311 pKa = 11.03SYY313 pKa = 10.74VYY315 pKa = 11.39SNMLFPSFSALSALGAAWAFPPGRR339 pKa = 11.84RR340 pKa = 11.84SVGRR344 pKa = 11.84GSFNRR349 pKa = 11.84HH350 pKa = 5.65TITGAGDD357 pKa = 3.53PQGKK361 pKa = 9.08KK362 pKa = 8.82WLTLVPKK369 pKa = 9.85KK370 pKa = 10.95VEE372 pKa = 4.33WITDD376 pKa = 3.41DD377 pKa = 3.76TMTQSEE383 pKa = 4.91IDD385 pKa = 3.45TTIATIYY392 pKa = 8.98VAQGAPKK399 pKa = 9.73ASGYY403 pKa = 10.5KK404 pKa = 9.78FQTFQGPLVDD414 pKa = 5.69DD415 pKa = 4.75PLSVQPWAVMRR426 pKa = 11.84VKK428 pKa = 10.67SVWQLGNLRR437 pKa = 11.84RR438 pKa = 11.84PYY440 pKa = 9.58PWDD443 pKa = 3.54VNWYY447 pKa = 9.18NQVADD452 pKa = 3.51WW453 pKa = 4.03
MM1 pKa = 6.6VRR3 pKa = 11.84YY4 pKa = 9.35RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84GVFYY16 pKa = 11.07SLGRR20 pKa = 11.84RR21 pKa = 11.84GWIRR25 pKa = 11.84HH26 pKa = 3.82RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84TWWGKK38 pKa = 8.44FRR40 pKa = 11.84HH41 pKa = 5.05ARR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84SFSQRR50 pKa = 11.84VKK52 pKa = 10.66RR53 pKa = 11.84RR54 pKa = 11.84FHH56 pKa = 6.18NPHH59 pKa = 6.89PGTYY63 pKa = 9.85LVRR66 pKa = 11.84LPRR69 pKa = 11.84PQTTLTIFFQGIVYY83 pKa = 9.57FPTAKK88 pKa = 9.68MYY90 pKa = 10.85ASRR93 pKa = 11.84NQLADD98 pKa = 3.63LNKK101 pKa = 9.23ITTCRR106 pKa = 11.84VGLMNINFRR115 pKa = 11.84EE116 pKa = 4.14FLLAALPLDD125 pKa = 4.57AYY127 pKa = 10.77SKK129 pKa = 10.87LGGPIVGPQYY139 pKa = 11.15VSGGGSSAGTHH150 pKa = 7.04DD151 pKa = 3.34EE152 pKa = 4.4TMWPFTARR160 pKa = 11.84PVTNKK165 pKa = 9.83QPGASPAEE173 pKa = 3.89WWRR176 pKa = 11.84WALLLMFPRR185 pKa = 11.84EE186 pKa = 3.78PNMFFRR192 pKa = 11.84QPQQPTLEE200 pKa = 4.06QMGEE204 pKa = 4.01IFGGWQLYY212 pKa = 8.37RR213 pKa = 11.84HH214 pKa = 6.77IKK216 pKa = 8.46TKK218 pKa = 9.5VAIMAAQDD226 pKa = 3.32GGAFSPVASLVSQNNYY242 pKa = 6.08FWRR245 pKa = 11.84RR246 pKa = 11.84QEE248 pKa = 3.83RR249 pKa = 11.84GTPKK253 pKa = 10.0TGEE256 pKa = 3.9PPMTGMIRR264 pKa = 11.84RR265 pKa = 11.84GTGSLEE271 pKa = 3.89SSKK274 pKa = 10.8PADD277 pKa = 3.74EE278 pKa = 4.79YY279 pKa = 11.38YY280 pKa = 10.3IPANPPNPPDD290 pKa = 3.38WPGEE294 pKa = 4.03SSAVIDD300 pKa = 3.91TPYY303 pKa = 10.56KK304 pKa = 9.35IAAVRR309 pKa = 11.84SKK311 pKa = 11.03SYY313 pKa = 10.74VYY315 pKa = 11.39SNMLFPSFSALSALGAAWAFPPGRR339 pKa = 11.84RR340 pKa = 11.84SVGRR344 pKa = 11.84GSFNRR349 pKa = 11.84HH350 pKa = 5.65TITGAGDD357 pKa = 3.53PQGKK361 pKa = 9.08KK362 pKa = 8.82WLTLVPKK369 pKa = 9.85KK370 pKa = 10.95VEE372 pKa = 4.33WITDD376 pKa = 3.41DD377 pKa = 3.76TMTQSEE383 pKa = 4.91IDD385 pKa = 3.45TTIATIYY392 pKa = 8.98VAQGAPKK399 pKa = 9.73ASGYY403 pKa = 10.5KK404 pKa = 9.78FQTFQGPLVDD414 pKa = 5.69DD415 pKa = 4.75PLSVQPWAVMRR426 pKa = 11.84VKK428 pKa = 10.67SVWQLGNLRR437 pKa = 11.84RR438 pKa = 11.84PYY440 pKa = 9.58PWDD443 pKa = 3.54VNWYY447 pKa = 9.18NQVADD452 pKa = 3.51WW453 pKa = 4.03
Molecular weight: 51.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
789 |
111 |
453 |
263.0 |
29.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.478 ± 0.476 | 1.394 ± 0.944 |
4.816 ± 1.359 | 4.563 ± 1.397 |
3.802 ± 1.045 | 8.745 ± 0.689 |
1.521 ± 0.145 | 3.676 ± 0.362 |
3.676 ± 0.417 | 6.337 ± 1.264 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.028 ± 0.659 | 3.295 ± 0.202 |
7.858 ± 0.942 | 4.056 ± 0.71 |
10.266 ± 0.103 | 7.605 ± 1.458 |
7.351 ± 1.265 | 5.577 ± 0.253 |
3.169 ± 0.839 | 2.788 ± 0.995 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
