
Avon-Heathcote Estuary associated circular virus 14
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
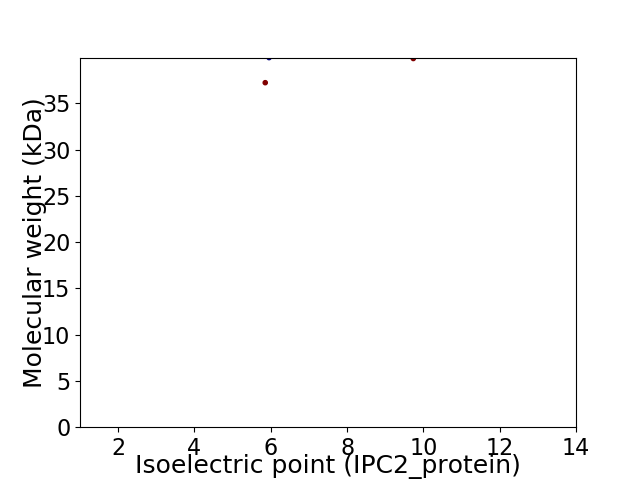
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMG6|A0A0C5IMG6_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 14 OX=1618237 PE=3 SV=1
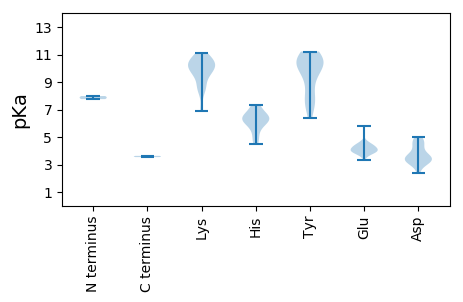
MM1 pKa = 7.76SGRR4 pKa = 11.84NWCFTINNPEE14 pKa = 3.94LTDD17 pKa = 4.26YY18 pKa = 10.95PEE20 pKa = 3.98QWPVTHH26 pKa = 7.33LKK28 pKa = 9.61TILYY32 pKa = 7.71QVEE35 pKa = 4.17MGEE38 pKa = 4.31EE39 pKa = 4.25EE40 pKa = 4.29TLHH43 pKa = 5.47LQGYY47 pKa = 8.95LEE49 pKa = 4.48IDD51 pKa = 3.57SPRR54 pKa = 11.84AIGWLKK60 pKa = 10.23GLNGRR65 pKa = 11.84AHH67 pKa = 6.06WEE69 pKa = 3.81RR70 pKa = 11.84RR71 pKa = 11.84KK72 pKa = 9.19GTRR75 pKa = 11.84SQAILYY81 pKa = 8.25CCKK84 pKa = 9.99EE85 pKa = 4.03DD86 pKa = 3.63SRR88 pKa = 11.84LVRR91 pKa = 11.84PRR93 pKa = 11.84LWTSRR98 pKa = 11.84DD99 pKa = 3.77LSWEE103 pKa = 4.0DD104 pKa = 4.62LDD106 pKa = 4.85PDD108 pKa = 4.87SLTSCLNSRR117 pKa = 11.84GITLATSEE125 pKa = 4.55NGNCGTNSRR134 pKa = 11.84LLEE137 pKa = 4.32IKK139 pKa = 10.06EE140 pKa = 4.12KK141 pKa = 10.67LSEE144 pKa = 4.01RR145 pKa = 11.84SSSIEE150 pKa = 3.96EE151 pKa = 3.91IADD154 pKa = 3.3EE155 pKa = 5.83HH156 pKa = 6.33FDD158 pKa = 3.08LWVRR162 pKa = 11.84HH163 pKa = 4.51YY164 pKa = 10.83RR165 pKa = 11.84AFEE168 pKa = 3.87KK169 pKa = 10.93YY170 pKa = 7.1MTMKK174 pKa = 10.78AEE176 pKa = 3.92PRR178 pKa = 11.84NWEE181 pKa = 4.24CEE183 pKa = 3.98VHH185 pKa = 6.37VLYY188 pKa = 11.02GPTGTGKK195 pKa = 10.57SKK197 pKa = 10.04WAMDD201 pKa = 4.67EE202 pKa = 4.16YY203 pKa = 10.71PGAYY207 pKa = 7.89WKK209 pKa = 10.09QRR211 pKa = 11.84SKK213 pKa = 9.66WWDD216 pKa = 3.52GYY218 pKa = 10.73VGHH221 pKa = 6.3EE222 pKa = 3.95TVIIDD227 pKa = 4.73EE228 pKa = 4.61YY229 pKa = 11.22YY230 pKa = 10.33GWLPFDD236 pKa = 4.48LLLRR240 pKa = 11.84LCDD243 pKa = 4.06RR244 pKa = 11.84YY245 pKa = 10.36PLLVEE250 pKa = 4.4SKK252 pKa = 10.39GGQLQFVAKK261 pKa = 9.59TIIFTTNKK269 pKa = 10.19CPDD272 pKa = 2.59QWYY275 pKa = 10.35NSDD278 pKa = 3.12SCYY281 pKa = 10.45FPAFEE286 pKa = 4.06RR287 pKa = 11.84RR288 pKa = 11.84VDD290 pKa = 3.03KK291 pKa = 10.34WHH293 pKa = 6.34YY294 pKa = 9.77LPSLGFHH301 pKa = 6.64SRR303 pKa = 11.84FTRR306 pKa = 11.84FKK308 pKa = 9.4EE309 pKa = 4.4FKK311 pKa = 10.39DD312 pKa = 3.62VTSS315 pKa = 3.63
MM1 pKa = 7.76SGRR4 pKa = 11.84NWCFTINNPEE14 pKa = 3.94LTDD17 pKa = 4.26YY18 pKa = 10.95PEE20 pKa = 3.98QWPVTHH26 pKa = 7.33LKK28 pKa = 9.61TILYY32 pKa = 7.71QVEE35 pKa = 4.17MGEE38 pKa = 4.31EE39 pKa = 4.25EE40 pKa = 4.29TLHH43 pKa = 5.47LQGYY47 pKa = 8.95LEE49 pKa = 4.48IDD51 pKa = 3.57SPRR54 pKa = 11.84AIGWLKK60 pKa = 10.23GLNGRR65 pKa = 11.84AHH67 pKa = 6.06WEE69 pKa = 3.81RR70 pKa = 11.84RR71 pKa = 11.84KK72 pKa = 9.19GTRR75 pKa = 11.84SQAILYY81 pKa = 8.25CCKK84 pKa = 9.99EE85 pKa = 4.03DD86 pKa = 3.63SRR88 pKa = 11.84LVRR91 pKa = 11.84PRR93 pKa = 11.84LWTSRR98 pKa = 11.84DD99 pKa = 3.77LSWEE103 pKa = 4.0DD104 pKa = 4.62LDD106 pKa = 4.85PDD108 pKa = 4.87SLTSCLNSRR117 pKa = 11.84GITLATSEE125 pKa = 4.55NGNCGTNSRR134 pKa = 11.84LLEE137 pKa = 4.32IKK139 pKa = 10.06EE140 pKa = 4.12KK141 pKa = 10.67LSEE144 pKa = 4.01RR145 pKa = 11.84SSSIEE150 pKa = 3.96EE151 pKa = 3.91IADD154 pKa = 3.3EE155 pKa = 5.83HH156 pKa = 6.33FDD158 pKa = 3.08LWVRR162 pKa = 11.84HH163 pKa = 4.51YY164 pKa = 10.83RR165 pKa = 11.84AFEE168 pKa = 3.87KK169 pKa = 10.93YY170 pKa = 7.1MTMKK174 pKa = 10.78AEE176 pKa = 3.92PRR178 pKa = 11.84NWEE181 pKa = 4.24CEE183 pKa = 3.98VHH185 pKa = 6.37VLYY188 pKa = 11.02GPTGTGKK195 pKa = 10.57SKK197 pKa = 10.04WAMDD201 pKa = 4.67EE202 pKa = 4.16YY203 pKa = 10.71PGAYY207 pKa = 7.89WKK209 pKa = 10.09QRR211 pKa = 11.84SKK213 pKa = 9.66WWDD216 pKa = 3.52GYY218 pKa = 10.73VGHH221 pKa = 6.3EE222 pKa = 3.95TVIIDD227 pKa = 4.73EE228 pKa = 4.61YY229 pKa = 11.22YY230 pKa = 10.33GWLPFDD236 pKa = 4.48LLLRR240 pKa = 11.84LCDD243 pKa = 4.06RR244 pKa = 11.84YY245 pKa = 10.36PLLVEE250 pKa = 4.4SKK252 pKa = 10.39GGQLQFVAKK261 pKa = 9.59TIIFTTNKK269 pKa = 10.19CPDD272 pKa = 2.59QWYY275 pKa = 10.35NSDD278 pKa = 3.12SCYY281 pKa = 10.45FPAFEE286 pKa = 4.06RR287 pKa = 11.84RR288 pKa = 11.84VDD290 pKa = 3.03KK291 pKa = 10.34WHH293 pKa = 6.34YY294 pKa = 9.77LPSLGFHH301 pKa = 6.64SRR303 pKa = 11.84FTRR306 pKa = 11.84FKK308 pKa = 9.4EE309 pKa = 4.4FKK311 pKa = 10.39DD312 pKa = 3.62VTSS315 pKa = 3.63
Molecular weight: 37.22 kDa
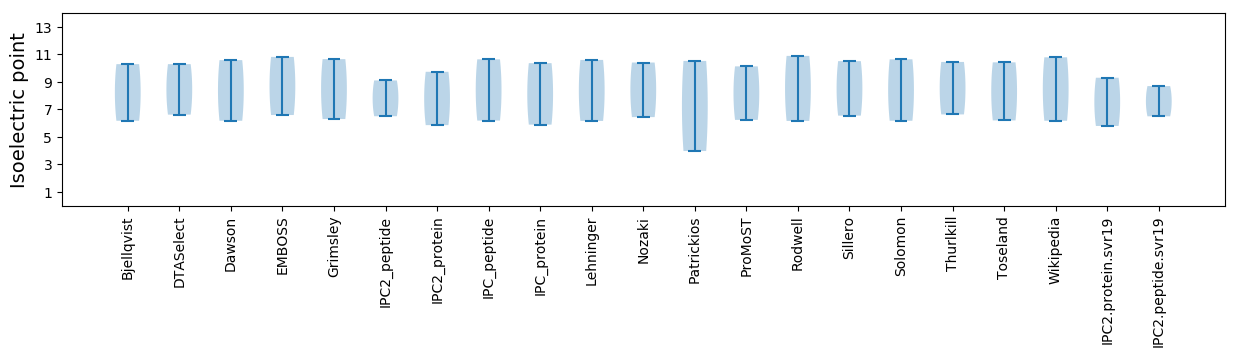
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMG6|A0A0C5IMG6_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 14 OX=1618237 PE=3 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84GRR4 pKa = 11.84KK5 pKa = 8.65RR6 pKa = 11.84DD7 pKa = 3.2HH8 pKa = 6.7RR9 pKa = 11.84GRR11 pKa = 11.84FIRR14 pKa = 11.84ATVVGAGRR22 pKa = 11.84AIRR25 pKa = 11.84YY26 pKa = 7.33AAGVGSGFAGLASSFARR43 pKa = 11.84TRR45 pKa = 11.84RR46 pKa = 11.84NRR48 pKa = 11.84MTSGQGVTFEE58 pKa = 4.13NDD60 pKa = 2.35RR61 pKa = 11.84TRR63 pKa = 11.84IYY65 pKa = 10.24RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.5RR69 pKa = 11.84APARR73 pKa = 11.84KK74 pKa = 8.29RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84WRR79 pKa = 11.84KK80 pKa = 8.57ILRR83 pKa = 11.84VNNAVDD89 pKa = 3.52QTKK92 pKa = 10.77LGTRR96 pKa = 11.84TILFNKK102 pKa = 8.77SQTYY106 pKa = 10.14GNSTAGQHH114 pKa = 4.94GLAWAALYY122 pKa = 9.36GANSANSVLNDD133 pKa = 4.22LRR135 pKa = 11.84AIWSNEE141 pKa = 3.32ALNQNPTAGGGPTSYY156 pKa = 11.2NSTKK160 pKa = 10.01MFFKK164 pKa = 10.75SAVLDD169 pKa = 3.43MTIRR173 pKa = 11.84NVSSVGDD180 pKa = 4.98DD181 pKa = 3.08ITSPADD187 pKa = 3.27VPLEE191 pKa = 3.74LDD193 pKa = 3.21IYY195 pKa = 11.0EE196 pKa = 4.35MSSKK200 pKa = 10.48CRR202 pKa = 11.84MVEE205 pKa = 3.7NDD207 pKa = 3.83AGTALKK213 pKa = 9.84QWNNIPEE220 pKa = 4.4TFDD223 pKa = 3.32EE224 pKa = 4.73ALDD227 pKa = 3.51NTFAIDD233 pKa = 4.35GAGTKK238 pKa = 10.1CDD240 pKa = 3.72LNLRR244 pKa = 11.84GVTPWEE250 pKa = 3.79NTFALSRR257 pKa = 11.84YY258 pKa = 8.89KK259 pKa = 10.94YY260 pKa = 10.07KK261 pKa = 10.17IHH263 pKa = 7.24KK264 pKa = 6.91KK265 pKa = 6.96TKK267 pKa = 8.48YY268 pKa = 10.06RR269 pKa = 11.84IGRR272 pKa = 11.84GNSITYY278 pKa = 7.76QFRR281 pKa = 11.84DD282 pKa = 3.53PKK284 pKa = 11.12NRR286 pKa = 11.84TTSHH290 pKa = 6.39NALGDD295 pKa = 3.49LAGFNKK301 pKa = 10.32PGWTHH306 pKa = 6.26NLLIVYY312 pKa = 9.55KK313 pKa = 9.7SVPGYY318 pKa = 10.69SVGTAPTDD326 pKa = 3.12IQEE329 pKa = 4.54RR330 pKa = 11.84IEE332 pKa = 3.66IGYY335 pKa = 6.38TRR337 pKa = 11.84KK338 pKa = 8.48YY339 pKa = 7.3TYY341 pKa = 10.18KK342 pKa = 10.6VEE344 pKa = 4.15GHH346 pKa = 5.9SDD348 pKa = 3.18ARR350 pKa = 11.84SLYY353 pKa = 10.16INSS356 pKa = 3.54
MM1 pKa = 7.98RR2 pKa = 11.84GRR4 pKa = 11.84KK5 pKa = 8.65RR6 pKa = 11.84DD7 pKa = 3.2HH8 pKa = 6.7RR9 pKa = 11.84GRR11 pKa = 11.84FIRR14 pKa = 11.84ATVVGAGRR22 pKa = 11.84AIRR25 pKa = 11.84YY26 pKa = 7.33AAGVGSGFAGLASSFARR43 pKa = 11.84TRR45 pKa = 11.84RR46 pKa = 11.84NRR48 pKa = 11.84MTSGQGVTFEE58 pKa = 4.13NDD60 pKa = 2.35RR61 pKa = 11.84TRR63 pKa = 11.84IYY65 pKa = 10.24RR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.5RR69 pKa = 11.84APARR73 pKa = 11.84KK74 pKa = 8.29RR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84WRR79 pKa = 11.84KK80 pKa = 8.57ILRR83 pKa = 11.84VNNAVDD89 pKa = 3.52QTKK92 pKa = 10.77LGTRR96 pKa = 11.84TILFNKK102 pKa = 8.77SQTYY106 pKa = 10.14GNSTAGQHH114 pKa = 4.94GLAWAALYY122 pKa = 9.36GANSANSVLNDD133 pKa = 4.22LRR135 pKa = 11.84AIWSNEE141 pKa = 3.32ALNQNPTAGGGPTSYY156 pKa = 11.2NSTKK160 pKa = 10.01MFFKK164 pKa = 10.75SAVLDD169 pKa = 3.43MTIRR173 pKa = 11.84NVSSVGDD180 pKa = 4.98DD181 pKa = 3.08ITSPADD187 pKa = 3.27VPLEE191 pKa = 3.74LDD193 pKa = 3.21IYY195 pKa = 11.0EE196 pKa = 4.35MSSKK200 pKa = 10.48CRR202 pKa = 11.84MVEE205 pKa = 3.7NDD207 pKa = 3.83AGTALKK213 pKa = 9.84QWNNIPEE220 pKa = 4.4TFDD223 pKa = 3.32EE224 pKa = 4.73ALDD227 pKa = 3.51NTFAIDD233 pKa = 4.35GAGTKK238 pKa = 10.1CDD240 pKa = 3.72LNLRR244 pKa = 11.84GVTPWEE250 pKa = 3.79NTFALSRR257 pKa = 11.84YY258 pKa = 8.89KK259 pKa = 10.94YY260 pKa = 10.07KK261 pKa = 10.17IHH263 pKa = 7.24KK264 pKa = 6.91KK265 pKa = 6.96TKK267 pKa = 8.48YY268 pKa = 10.06RR269 pKa = 11.84IGRR272 pKa = 11.84GNSITYY278 pKa = 7.76QFRR281 pKa = 11.84DD282 pKa = 3.53PKK284 pKa = 11.12NRR286 pKa = 11.84TTSHH290 pKa = 6.39NALGDD295 pKa = 3.49LAGFNKK301 pKa = 10.32PGWTHH306 pKa = 6.26NLLIVYY312 pKa = 9.55KK313 pKa = 9.7SVPGYY318 pKa = 10.69SVGTAPTDD326 pKa = 3.12IQEE329 pKa = 4.54RR330 pKa = 11.84IEE332 pKa = 3.66IGYY335 pKa = 6.38TRR337 pKa = 11.84KK338 pKa = 8.48YY339 pKa = 7.3TYY341 pKa = 10.18KK342 pKa = 10.6VEE344 pKa = 4.15GHH346 pKa = 5.9SDD348 pKa = 3.18ARR350 pKa = 11.84SLYY353 pKa = 10.16INSS356 pKa = 3.54
Molecular weight: 39.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
671 |
315 |
356 |
335.5 |
38.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.408 ± 1.876 | 1.639 ± 0.783 |
5.365 ± 0.225 | 5.663 ± 1.871 |
3.577 ± 0.15 | 7.75 ± 0.901 |
2.235 ± 0.4 | 4.769 ± 0.413 |
5.812 ± 0.063 | 7.899 ± 1.25 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.639 ± 0.033 | 5.514 ± 1.301 |
3.726 ± 0.462 | 2.385 ± 0.1 |
8.793 ± 0.959 | 7.154 ± 0.095 |
7.452 ± 0.709 | 4.322 ± 0.33 |
3.13 ± 1.05 | 4.769 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
