
Marinobacter persicus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
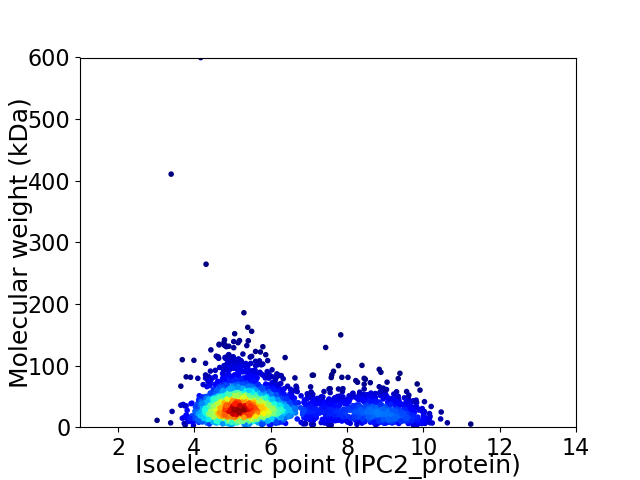
Virtual 2D-PAGE plot for 2913 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3UGW0|A0A1I3UGW0_9ALTE Secondary thiamine-phosphate synthase enzyme OS=Marinobacter persicus OX=930118 GN=SAMN05216429_106106 PE=3 SV=1
MM1 pKa = 7.39SSSGGGDD8 pKa = 3.1INIGGGSGLYY18 pKa = 9.35TVDD21 pKa = 3.0GCAYY25 pKa = 10.37SCGSNPAKK33 pKa = 9.88WDD35 pKa = 3.51GNHH38 pKa = 6.93TYY40 pKa = 10.79GQTLEE45 pKa = 4.22CKK47 pKa = 10.2GLGVTPEE54 pKa = 3.71EE55 pKa = 4.59AEE57 pKa = 4.07AGEE60 pKa = 4.98AGPADD65 pKa = 4.63LNCATDD71 pKa = 3.36SSGKK75 pKa = 9.55EE76 pKa = 3.68ICSDD80 pKa = 3.91PDD82 pKa = 3.35NPSCVTIDD90 pKa = 4.92GISTCPSEE98 pKa = 5.34GAICDD103 pKa = 4.19EE104 pKa = 4.49LNGDD108 pKa = 5.04LGCIDD113 pKa = 4.16PAEE116 pKa = 4.41EE117 pKa = 4.3GCAYY121 pKa = 10.78RR122 pKa = 11.84NGKK125 pKa = 8.47KK126 pKa = 9.45EE127 pKa = 4.02CYY129 pKa = 9.9DD130 pKa = 3.03IFGQYY135 pKa = 9.11IEE137 pKa = 5.38HH138 pKa = 7.9DD139 pKa = 4.4SPDD142 pKa = 3.48HH143 pKa = 6.48PNNGGNMDD151 pKa = 4.32GDD153 pKa = 4.19TSNDD157 pKa = 3.21MTDD160 pKa = 3.5PRR162 pKa = 11.84PEE164 pKa = 4.68GEE166 pKa = 4.99GGDD169 pKa = 4.4PNNQPGDD176 pKa = 3.75STGEE180 pKa = 3.97GGEE183 pKa = 4.19ATEE186 pKa = 4.48GTAKK190 pKa = 10.47EE191 pKa = 4.16SLEE194 pKa = 3.81QQKK197 pKa = 10.34IGNEE201 pKa = 3.87KK202 pKa = 10.37LDD204 pKa = 4.59GIEE207 pKa = 4.15EE208 pKa = 4.35STGNIDD214 pKa = 3.65EE215 pKa = 5.44AIEE218 pKa = 4.13AALDD222 pKa = 3.47ASRR225 pKa = 11.84GDD227 pKa = 3.49GTALKK232 pKa = 10.72NGIDD236 pKa = 3.98DD237 pKa = 5.57AISNAGDD244 pKa = 3.78SAVGDD249 pKa = 4.1LDD251 pKa = 3.63EE252 pKa = 6.69HH253 pKa = 8.03IDD255 pKa = 4.9GIDD258 pKa = 3.32SPAPMNEE265 pKa = 3.68GDD267 pKa = 5.11LGAIEE272 pKa = 5.0NNVTNLFAGSMQCRR286 pKa = 11.84DD287 pKa = 3.58LVFGTDD293 pKa = 3.53EE294 pKa = 3.55ISYY297 pKa = 9.25TITCNDD303 pKa = 2.87MSMIRR308 pKa = 11.84DD309 pKa = 3.42MLGFLLYY316 pKa = 10.39GFTVIRR322 pKa = 11.84LFNVALRR329 pKa = 11.84PAAKK333 pKa = 8.27GTPP336 pKa = 3.43
MM1 pKa = 7.39SSSGGGDD8 pKa = 3.1INIGGGSGLYY18 pKa = 9.35TVDD21 pKa = 3.0GCAYY25 pKa = 10.37SCGSNPAKK33 pKa = 9.88WDD35 pKa = 3.51GNHH38 pKa = 6.93TYY40 pKa = 10.79GQTLEE45 pKa = 4.22CKK47 pKa = 10.2GLGVTPEE54 pKa = 3.71EE55 pKa = 4.59AEE57 pKa = 4.07AGEE60 pKa = 4.98AGPADD65 pKa = 4.63LNCATDD71 pKa = 3.36SSGKK75 pKa = 9.55EE76 pKa = 3.68ICSDD80 pKa = 3.91PDD82 pKa = 3.35NPSCVTIDD90 pKa = 4.92GISTCPSEE98 pKa = 5.34GAICDD103 pKa = 4.19EE104 pKa = 4.49LNGDD108 pKa = 5.04LGCIDD113 pKa = 4.16PAEE116 pKa = 4.41EE117 pKa = 4.3GCAYY121 pKa = 10.78RR122 pKa = 11.84NGKK125 pKa = 8.47KK126 pKa = 9.45EE127 pKa = 4.02CYY129 pKa = 9.9DD130 pKa = 3.03IFGQYY135 pKa = 9.11IEE137 pKa = 5.38HH138 pKa = 7.9DD139 pKa = 4.4SPDD142 pKa = 3.48HH143 pKa = 6.48PNNGGNMDD151 pKa = 4.32GDD153 pKa = 4.19TSNDD157 pKa = 3.21MTDD160 pKa = 3.5PRR162 pKa = 11.84PEE164 pKa = 4.68GEE166 pKa = 4.99GGDD169 pKa = 4.4PNNQPGDD176 pKa = 3.75STGEE180 pKa = 3.97GGEE183 pKa = 4.19ATEE186 pKa = 4.48GTAKK190 pKa = 10.47EE191 pKa = 4.16SLEE194 pKa = 3.81QQKK197 pKa = 10.34IGNEE201 pKa = 3.87KK202 pKa = 10.37LDD204 pKa = 4.59GIEE207 pKa = 4.15EE208 pKa = 4.35STGNIDD214 pKa = 3.65EE215 pKa = 5.44AIEE218 pKa = 4.13AALDD222 pKa = 3.47ASRR225 pKa = 11.84GDD227 pKa = 3.49GTALKK232 pKa = 10.72NGIDD236 pKa = 3.98DD237 pKa = 5.57AISNAGDD244 pKa = 3.78SAVGDD249 pKa = 4.1LDD251 pKa = 3.63EE252 pKa = 6.69HH253 pKa = 8.03IDD255 pKa = 4.9GIDD258 pKa = 3.32SPAPMNEE265 pKa = 3.68GDD267 pKa = 5.11LGAIEE272 pKa = 5.0NNVTNLFAGSMQCRR286 pKa = 11.84DD287 pKa = 3.58LVFGTDD293 pKa = 3.53EE294 pKa = 3.55ISYY297 pKa = 9.25TITCNDD303 pKa = 2.87MSMIRR308 pKa = 11.84DD309 pKa = 3.42MLGFLLYY316 pKa = 10.39GFTVIRR322 pKa = 11.84LFNVALRR329 pKa = 11.84PAAKK333 pKa = 8.27GTPP336 pKa = 3.43
Molecular weight: 34.75 kDa
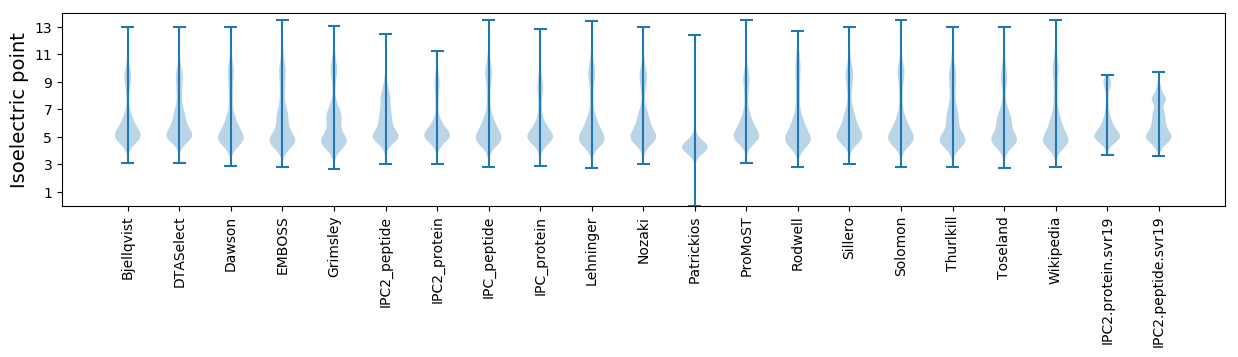
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3V6M6|A0A1I3V6M6_9ALTE RecBCD enzyme subunit RecD OS=Marinobacter persicus OX=930118 GN=recD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.91VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.44GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84AHH16 pKa = 6.12GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.91VLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.44GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
959929 |
29 |
5842 |
329.5 |
36.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.819 ± 0.05 | 0.902 ± 0.015 |
5.87 ± 0.045 | 6.904 ± 0.05 |
3.687 ± 0.029 | 7.78 ± 0.061 |
2.26 ± 0.025 | 5.124 ± 0.035 |
3.758 ± 0.043 | 10.704 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.556 ± 0.021 | 3.293 ± 0.037 |
4.673 ± 0.036 | 4.246 ± 0.033 |
6.524 ± 0.039 | 5.647 ± 0.038 |
5.132 ± 0.035 | 7.256 ± 0.041 |
1.357 ± 0.017 | 2.509 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
