
Paraburkholderia sp. PDC91
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Paraburkholderia; unclassified Paraburkholderia
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
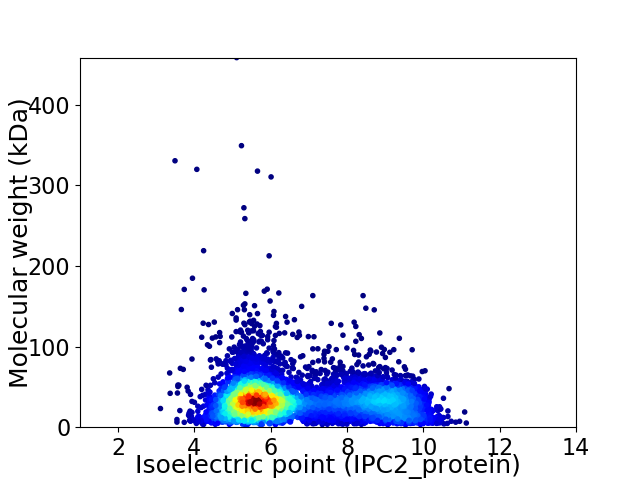
Virtual 2D-PAGE plot for 6876 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W7FHD9|A0A2W7FHD9_9BURK Amino acid ABC transporter substrate-binding protein (PAAT family) OS=Paraburkholderia sp. PDC91 OX=2135498 GN=C7514_116141 PE=4 SV=1
MM1 pKa = 7.36NKK3 pKa = 9.75RR4 pKa = 11.84YY5 pKa = 9.72RR6 pKa = 11.84KK7 pKa = 9.32QPMRR11 pKa = 11.84TRR13 pKa = 11.84TIASVVAALFEE24 pKa = 4.26ASYY27 pKa = 11.27ASSLLANPTGAQVVAGSVNVNSPTAGQMNITQGTQNAIVNWNTFSIGAGEE77 pKa = 4.68SVTVAQPSASAALLNRR93 pKa = 11.84VVGNDD98 pKa = 3.18PSTIAGRR105 pKa = 11.84LQANGKK111 pKa = 9.22VFLVNPAGVIFSPGSSVNVGSLVASTLNISDD142 pKa = 4.24ADD144 pKa = 3.86FLAGNYY150 pKa = 9.46HH151 pKa = 6.54FVGASPAGVSNAGTLAAQQGGTIALLGGTVSNSGTVTAKK190 pKa = 10.71LGTVALGAGSDD201 pKa = 3.33ISLDD205 pKa = 3.66FAGDD209 pKa = 3.44GLTTLTINSAAAQALVGNTGTLAADD234 pKa = 3.98GGLVVMSAQTADD246 pKa = 3.49ALASSVVNQQGIVRR260 pKa = 11.84AQSLAEE266 pKa = 4.06RR267 pKa = 11.84NGHH270 pKa = 5.83IVLDD274 pKa = 4.29GGSNGTTTVSGTLDD288 pKa = 3.26ATGGAGLSGGRR299 pKa = 11.84IDD301 pKa = 3.52ATGRR305 pKa = 11.84DD306 pKa = 3.62VALLAGANVDD316 pKa = 3.25ASGAAGGGTVNLGGGAGGQDD336 pKa = 3.31PAIRR340 pKa = 11.84NADD343 pKa = 3.82TLSIDD348 pKa = 3.71PAAQVHH354 pKa = 6.79ADD356 pKa = 3.38ALTNGNGGSVSAYY369 pKa = 10.41SDD371 pKa = 3.45TSGLLYY377 pKa = 9.52GTLTAKK383 pKa = 10.57GGPQGGNGGMVEE395 pKa = 4.32TSSHH399 pKa = 5.69YY400 pKa = 10.98LDD402 pKa = 5.17LLGSTIDD409 pKa = 3.53VSAPAGNGGMWKK421 pKa = 9.47IDD423 pKa = 3.87PYY425 pKa = 11.54EE426 pKa = 4.03MNITDD431 pKa = 4.33NPNEE435 pKa = 4.15TVPTGGSLVLASEE448 pKa = 4.96IEE450 pKa = 3.9QSLNQGARR458 pKa = 11.84VIVDD462 pKa = 3.85AASTDD467 pKa = 3.51PGSINVEE474 pKa = 4.3SPISMTGARR483 pKa = 11.84PASLTLMASGSISVEE498 pKa = 3.63PNANITSTGGALDD511 pKa = 3.78LTLNAAIGADD521 pKa = 3.62PTAGSVSINAATIDD535 pKa = 3.9TNGGAFTINAANSGGQSPVSIMDD558 pKa = 3.52STISTGAGAITINGNATTGGAIALGYY584 pKa = 7.93NTLLATTTGNVTLTGVAQQGDD605 pKa = 4.0GVDD608 pKa = 3.6IEE610 pKa = 4.61SSAVQTANGAIQISGAGTAFEE631 pKa = 3.99PTAYY635 pKa = 9.82GVYY638 pKa = 9.29ATGGYY643 pKa = 7.86TPAGSDD649 pKa = 3.35TASVPVLGATGTGSISVTGTATGNSATGVEE679 pKa = 4.28LDD681 pKa = 3.63GATVSAASGGVTINGTANAANVGNGVSLSDD711 pKa = 3.78AAIDD715 pKa = 3.71TASGNVSITGTATNQQVAVGYY736 pKa = 10.46ALSTTANGIALGYY749 pKa = 10.26SSVASIDD756 pKa = 3.25GDD758 pKa = 3.86LALNGSTQVMGDD770 pKa = 3.75GVRR773 pKa = 11.84MLQATLQTTGSGSIAVTGNANVPDD797 pKa = 4.01STALQTGVDD806 pKa = 3.81VAGTTVSASGTGSIALTGNANGEE829 pKa = 4.23SSDD832 pKa = 3.63GVVVIDD838 pKa = 3.82ATILGAAGAISVNGSATATANGGNAAGVAVQEE870 pKa = 4.43SALNSTTGAINVTGAALAGADD891 pKa = 3.43GLTYY895 pKa = 10.79SGDD898 pKa = 3.75GVTMYY903 pKa = 10.21YY904 pKa = 11.04GSIGTTSGNISVTGTATGANSGASFSNADD933 pKa = 3.2GVAVQSSYY941 pKa = 10.92GQQSAGTAGMAFSSTAGGSIDD962 pKa = 4.59IEE964 pKa = 4.66GSAQGQYY971 pKa = 10.95VIGVGLDD978 pKa = 3.28SAGISATTGPITVHH992 pKa = 6.35GSADD996 pKa = 3.57TTISGEE1002 pKa = 4.5GVDD1005 pKa = 3.96TDD1007 pKa = 3.49IALLSSTSGAITVNGSANASDD1028 pKa = 3.85AGTGRR1033 pKa = 11.84GVEE1036 pKa = 4.04LGYY1039 pKa = 9.68TSLTTTSGGVNVTGTVTGTPGQATGATIGAGYY1071 pKa = 8.27TPQQEE1076 pKa = 4.58EE1077 pKa = 4.13EE1078 pKa = 4.36GGPLTEE1084 pKa = 5.37AVAGDD1089 pKa = 4.19PPSVQTDD1096 pKa = 3.27SGNITIAGSAPATAQGSNALSLFSASIVSNSGAIGLTGTTTGTPTPGTSSTGVLLNGSNGDD1157 pKa = 3.45GFTNTSITTGSGALTIEE1174 pKa = 4.78GSGSGQLAAGVAIANATTITSTSGGAIDD1202 pKa = 3.64IRR1204 pKa = 11.84GVATSPTTSTAGGNIQYY1221 pKa = 10.7DD1222 pKa = 4.19YY1223 pKa = 10.25GTLIEE1228 pKa = 5.51DD1229 pKa = 4.28GSITSTMAGSTISIAGSTNTSDD1251 pKa = 3.14AGVAIGAVPIPSDD1264 pKa = 3.15QSVFFNTPYY1273 pKa = 9.35EE1274 pKa = 4.28TGPVTISGAPGGTLTLRR1291 pKa = 11.84AANDD1295 pKa = 3.4NTSQSLLARR1304 pKa = 11.84GATITGGGGTLAISSASVDD1323 pKa = 3.4PATFALSSQDD1333 pKa = 3.58APPITLFGTSGGMSIDD1349 pKa = 3.41AQTFAAFSQFGSLTLGSSTQTGLITVNGQCASASSSCQPVKK1390 pKa = 9.67PTLDD1394 pKa = 3.47MNLTLANAGAGSQGIQLPYY1413 pKa = 10.63GLDD1416 pKa = 3.49VGNHH1420 pKa = 4.74TLTLQSAGNVTDD1432 pKa = 4.24PGGIDD1437 pKa = 3.29AAQLVLAGPGTFTLTDD1453 pKa = 3.56PQNNVGEE1460 pKa = 4.39IAFEE1464 pKa = 3.88NAGNVDD1470 pKa = 4.01FSNPVGFVIGGGNSTLTGNLVAQATTGNISLGSTAAGGPTTNLSAGGTIDD1520 pKa = 4.94LVMEE1524 pKa = 4.43NGVFVSPDD1532 pKa = 3.18GGTVSATNGWRR1543 pKa = 11.84IWASTWSGEE1552 pKa = 4.04TRR1554 pKa = 11.84GNVQPNTSQPNFYY1567 pKa = 10.66GCTFGAGCSWGGTVPSSGDD1586 pKa = 3.08HH1587 pKa = 4.97YY1588 pKa = 11.35VYY1590 pKa = 10.63VARR1593 pKa = 11.84PTVTVTADD1601 pKa = 2.95GEE1603 pKa = 4.7TRR1605 pKa = 11.84VVGAPDD1611 pKa = 3.43PAFTYY1616 pKa = 10.45AVSGLINGDD1625 pKa = 3.41TAAGTLSGTLTSPATPGSPAGRR1647 pKa = 11.84YY1648 pKa = 9.86AIDD1651 pKa = 3.48PAFLSSVGYY1660 pKa = 9.87IVNDD1664 pKa = 3.3VPGTLTVTAAPTSTQSLAPRR1684 pKa = 11.84FAEE1687 pKa = 4.31DD1688 pKa = 4.4LPMQVGAQQSYY1699 pKa = 10.03FGNEE1703 pKa = 3.62EE1704 pKa = 3.69KK1705 pKa = 10.55TFVYY1709 pKa = 10.29EE1710 pKa = 4.73NNLQGTNICVGSSEE1724 pKa = 4.46PLFTTAPPGDD1734 pKa = 4.16NQDD1737 pKa = 3.77LLAVEE1742 pKa = 4.47WRR1744 pKa = 11.84RR1745 pKa = 11.84VRR1747 pKa = 11.84SQPNLNSCLLLNSQHH1762 pKa = 6.21GCGDD1766 pKa = 3.83FF1767 pKa = 3.65
MM1 pKa = 7.36NKK3 pKa = 9.75RR4 pKa = 11.84YY5 pKa = 9.72RR6 pKa = 11.84KK7 pKa = 9.32QPMRR11 pKa = 11.84TRR13 pKa = 11.84TIASVVAALFEE24 pKa = 4.26ASYY27 pKa = 11.27ASSLLANPTGAQVVAGSVNVNSPTAGQMNITQGTQNAIVNWNTFSIGAGEE77 pKa = 4.68SVTVAQPSASAALLNRR93 pKa = 11.84VVGNDD98 pKa = 3.18PSTIAGRR105 pKa = 11.84LQANGKK111 pKa = 9.22VFLVNPAGVIFSPGSSVNVGSLVASTLNISDD142 pKa = 4.24ADD144 pKa = 3.86FLAGNYY150 pKa = 9.46HH151 pKa = 6.54FVGASPAGVSNAGTLAAQQGGTIALLGGTVSNSGTVTAKK190 pKa = 10.71LGTVALGAGSDD201 pKa = 3.33ISLDD205 pKa = 3.66FAGDD209 pKa = 3.44GLTTLTINSAAAQALVGNTGTLAADD234 pKa = 3.98GGLVVMSAQTADD246 pKa = 3.49ALASSVVNQQGIVRR260 pKa = 11.84AQSLAEE266 pKa = 4.06RR267 pKa = 11.84NGHH270 pKa = 5.83IVLDD274 pKa = 4.29GGSNGTTTVSGTLDD288 pKa = 3.26ATGGAGLSGGRR299 pKa = 11.84IDD301 pKa = 3.52ATGRR305 pKa = 11.84DD306 pKa = 3.62VALLAGANVDD316 pKa = 3.25ASGAAGGGTVNLGGGAGGQDD336 pKa = 3.31PAIRR340 pKa = 11.84NADD343 pKa = 3.82TLSIDD348 pKa = 3.71PAAQVHH354 pKa = 6.79ADD356 pKa = 3.38ALTNGNGGSVSAYY369 pKa = 10.41SDD371 pKa = 3.45TSGLLYY377 pKa = 9.52GTLTAKK383 pKa = 10.57GGPQGGNGGMVEE395 pKa = 4.32TSSHH399 pKa = 5.69YY400 pKa = 10.98LDD402 pKa = 5.17LLGSTIDD409 pKa = 3.53VSAPAGNGGMWKK421 pKa = 9.47IDD423 pKa = 3.87PYY425 pKa = 11.54EE426 pKa = 4.03MNITDD431 pKa = 4.33NPNEE435 pKa = 4.15TVPTGGSLVLASEE448 pKa = 4.96IEE450 pKa = 3.9QSLNQGARR458 pKa = 11.84VIVDD462 pKa = 3.85AASTDD467 pKa = 3.51PGSINVEE474 pKa = 4.3SPISMTGARR483 pKa = 11.84PASLTLMASGSISVEE498 pKa = 3.63PNANITSTGGALDD511 pKa = 3.78LTLNAAIGADD521 pKa = 3.62PTAGSVSINAATIDD535 pKa = 3.9TNGGAFTINAANSGGQSPVSIMDD558 pKa = 3.52STISTGAGAITINGNATTGGAIALGYY584 pKa = 7.93NTLLATTTGNVTLTGVAQQGDD605 pKa = 4.0GVDD608 pKa = 3.6IEE610 pKa = 4.61SSAVQTANGAIQISGAGTAFEE631 pKa = 3.99PTAYY635 pKa = 9.82GVYY638 pKa = 9.29ATGGYY643 pKa = 7.86TPAGSDD649 pKa = 3.35TASVPVLGATGTGSISVTGTATGNSATGVEE679 pKa = 4.28LDD681 pKa = 3.63GATVSAASGGVTINGTANAANVGNGVSLSDD711 pKa = 3.78AAIDD715 pKa = 3.71TASGNVSITGTATNQQVAVGYY736 pKa = 10.46ALSTTANGIALGYY749 pKa = 10.26SSVASIDD756 pKa = 3.25GDD758 pKa = 3.86LALNGSTQVMGDD770 pKa = 3.75GVRR773 pKa = 11.84MLQATLQTTGSGSIAVTGNANVPDD797 pKa = 4.01STALQTGVDD806 pKa = 3.81VAGTTVSASGTGSIALTGNANGEE829 pKa = 4.23SSDD832 pKa = 3.63GVVVIDD838 pKa = 3.82ATILGAAGAISVNGSATATANGGNAAGVAVQEE870 pKa = 4.43SALNSTTGAINVTGAALAGADD891 pKa = 3.43GLTYY895 pKa = 10.79SGDD898 pKa = 3.75GVTMYY903 pKa = 10.21YY904 pKa = 11.04GSIGTTSGNISVTGTATGANSGASFSNADD933 pKa = 3.2GVAVQSSYY941 pKa = 10.92GQQSAGTAGMAFSSTAGGSIDD962 pKa = 4.59IEE964 pKa = 4.66GSAQGQYY971 pKa = 10.95VIGVGLDD978 pKa = 3.28SAGISATTGPITVHH992 pKa = 6.35GSADD996 pKa = 3.57TTISGEE1002 pKa = 4.5GVDD1005 pKa = 3.96TDD1007 pKa = 3.49IALLSSTSGAITVNGSANASDD1028 pKa = 3.85AGTGRR1033 pKa = 11.84GVEE1036 pKa = 4.04LGYY1039 pKa = 9.68TSLTTTSGGVNVTGTVTGTPGQATGATIGAGYY1071 pKa = 8.27TPQQEE1076 pKa = 4.58EE1077 pKa = 4.13EE1078 pKa = 4.36GGPLTEE1084 pKa = 5.37AVAGDD1089 pKa = 4.19PPSVQTDD1096 pKa = 3.27SGNITIAGSAPATAQGSNALSLFSASIVSNSGAIGLTGTTTGTPTPGTSSTGVLLNGSNGDD1157 pKa = 3.45GFTNTSITTGSGALTIEE1174 pKa = 4.78GSGSGQLAAGVAIANATTITSTSGGAIDD1202 pKa = 3.64IRR1204 pKa = 11.84GVATSPTTSTAGGNIQYY1221 pKa = 10.7DD1222 pKa = 4.19YY1223 pKa = 10.25GTLIEE1228 pKa = 5.51DD1229 pKa = 4.28GSITSTMAGSTISIAGSTNTSDD1251 pKa = 3.14AGVAIGAVPIPSDD1264 pKa = 3.15QSVFFNTPYY1273 pKa = 9.35EE1274 pKa = 4.28TGPVTISGAPGGTLTLRR1291 pKa = 11.84AANDD1295 pKa = 3.4NTSQSLLARR1304 pKa = 11.84GATITGGGGTLAISSASVDD1323 pKa = 3.4PATFALSSQDD1333 pKa = 3.58APPITLFGTSGGMSIDD1349 pKa = 3.41AQTFAAFSQFGSLTLGSSTQTGLITVNGQCASASSSCQPVKK1390 pKa = 9.67PTLDD1394 pKa = 3.47MNLTLANAGAGSQGIQLPYY1413 pKa = 10.63GLDD1416 pKa = 3.49VGNHH1420 pKa = 4.74TLTLQSAGNVTDD1432 pKa = 4.24PGGIDD1437 pKa = 3.29AAQLVLAGPGTFTLTDD1453 pKa = 3.56PQNNVGEE1460 pKa = 4.39IAFEE1464 pKa = 3.88NAGNVDD1470 pKa = 4.01FSNPVGFVIGGGNSTLTGNLVAQATTGNISLGSTAAGGPTTNLSAGGTIDD1520 pKa = 4.94LVMEE1524 pKa = 4.43NGVFVSPDD1532 pKa = 3.18GGTVSATNGWRR1543 pKa = 11.84IWASTWSGEE1552 pKa = 4.04TRR1554 pKa = 11.84GNVQPNTSQPNFYY1567 pKa = 10.66GCTFGAGCSWGGTVPSSGDD1586 pKa = 3.08HH1587 pKa = 4.97YY1588 pKa = 11.35VYY1590 pKa = 10.63VARR1593 pKa = 11.84PTVTVTADD1601 pKa = 2.95GEE1603 pKa = 4.7TRR1605 pKa = 11.84VVGAPDD1611 pKa = 3.43PAFTYY1616 pKa = 10.45AVSGLINGDD1625 pKa = 3.41TAAGTLSGTLTSPATPGSPAGRR1647 pKa = 11.84YY1648 pKa = 9.86AIDD1651 pKa = 3.48PAFLSSVGYY1660 pKa = 9.87IVNDD1664 pKa = 3.3VPGTLTVTAAPTSTQSLAPRR1684 pKa = 11.84FAEE1687 pKa = 4.31DD1688 pKa = 4.4LPMQVGAQQSYY1699 pKa = 10.03FGNEE1703 pKa = 3.62EE1704 pKa = 3.69KK1705 pKa = 10.55TFVYY1709 pKa = 10.29EE1710 pKa = 4.73NNLQGTNICVGSSEE1724 pKa = 4.46PLFTTAPPGDD1734 pKa = 4.16NQDD1737 pKa = 3.77LLAVEE1742 pKa = 4.47WRR1744 pKa = 11.84RR1745 pKa = 11.84VRR1747 pKa = 11.84SQPNLNSCLLLNSQHH1762 pKa = 6.21GCGDD1766 pKa = 3.83FF1767 pKa = 3.65
Molecular weight: 170.92 kDa
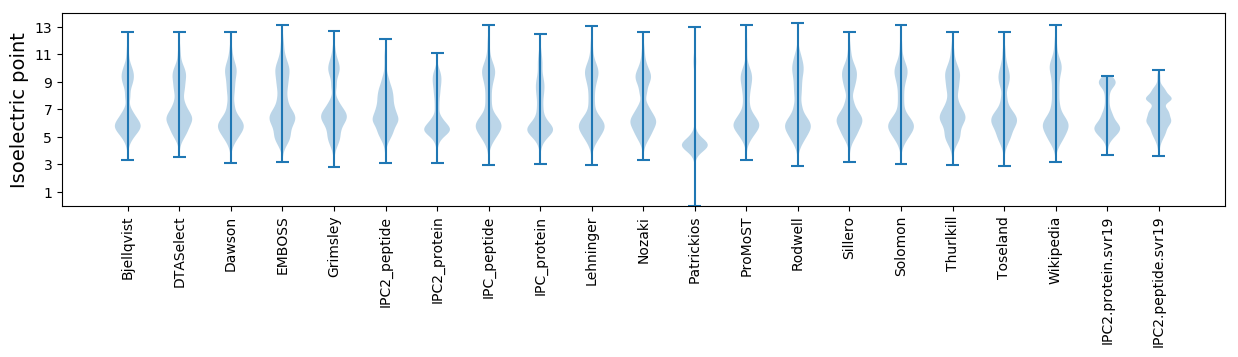
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W7GVK9|A0A2W7GVK9_9BURK 2Fe-2S ferredoxin OS=Paraburkholderia sp. PDC91 OX=2135498 GN=C7514_101623 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.06QPSVTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.0RR14 pKa = 11.84THH16 pKa = 5.76GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 8.74TAGGRR28 pKa = 11.84KK29 pKa = 9.04VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2242271 |
29 |
4351 |
326.1 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.978 ± 0.05 | 0.906 ± 0.009 |
5.599 ± 0.025 | 4.862 ± 0.034 |
3.633 ± 0.022 | 8.478 ± 0.041 |
2.321 ± 0.015 | 4.648 ± 0.023 |
2.672 ± 0.026 | 10.173 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.013 | 2.699 ± 0.022 |
5.247 ± 0.023 | 3.477 ± 0.021 |
7.208 ± 0.037 | 5.608 ± 0.026 |
5.471 ± 0.035 | 7.934 ± 0.024 |
1.375 ± 0.013 | 2.424 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
