
Limnohabitans sp. 2KL-17
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Limnohabitans; unclassified Limnohabitans
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
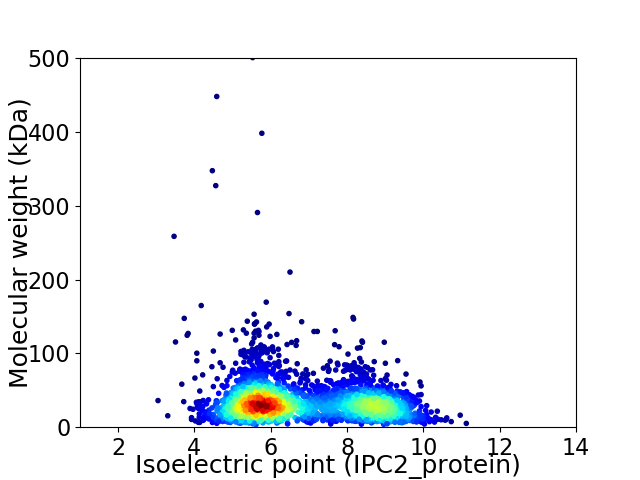
Virtual 2D-PAGE plot for 3184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A315DSE8|A0A315DSE8_9BURK Resolvase OS=Limnohabitans sp. 2KL-17 OX=1100704 GN=B9Z45_16290 PE=4 SV=1
MM1 pKa = 8.09VEE3 pKa = 3.96VSAIAITNLSDD14 pKa = 3.82SSVAEE19 pKa = 3.85NAAYY23 pKa = 8.96TSSTPSLSGTPIGAVTYY40 pKa = 10.46SLGGADD46 pKa = 3.71SALFTLNTGTGVVSMVARR64 pKa = 11.84NFEE67 pKa = 4.21SPDD70 pKa = 3.3DD71 pKa = 4.17AGANNVYY78 pKa = 10.61NYY80 pKa = 9.95TLTATDD86 pKa = 3.97ADD88 pKa = 4.47GNSADD93 pKa = 3.68QAVVLSVVDD102 pKa = 4.45FNDD105 pKa = 3.24NAPVFSSGATGSVPEE120 pKa = 4.24NAATTRR126 pKa = 11.84EE127 pKa = 4.34IYY129 pKa = 8.88TASTTDD135 pKa = 3.47ADD137 pKa = 3.63GTAANRR143 pKa = 11.84NVVYY147 pKa = 10.23SLKK150 pKa = 10.8AATGDD155 pKa = 3.51IALLDD160 pKa = 3.76IDD162 pKa = 4.22SATGKK167 pKa = 8.53VTLKK171 pKa = 10.91NSANYY176 pKa = 7.53EE177 pKa = 4.18AKK179 pKa = 10.12SSYY182 pKa = 10.87SFTVVATNVATSGTLSTEE200 pKa = 3.73QAVIANVTNVNDD212 pKa = 3.91APTVSAALASNASEE226 pKa = 4.33GDD228 pKa = 3.01SSYY231 pKa = 11.17TVNLLQGATDD241 pKa = 3.71ADD243 pKa = 3.78AGEE246 pKa = 4.3TATLSTQSLTYY257 pKa = 9.86QVNGGTASATLPAGVSITGNTLTVDD282 pKa = 3.61PANAVFNSLAFGQSQVIVASYY303 pKa = 10.66QVTDD307 pKa = 3.49VQGASVNQTATLTISGTNDD326 pKa = 3.1APTLSANTQTASYY339 pKa = 10.61ANDD342 pKa = 3.06ATAQTVFSGAFISAVDD358 pKa = 3.62AGQTIKK364 pKa = 10.88GLKK367 pKa = 8.54FTVSGITDD375 pKa = 3.75ASSEE379 pKa = 4.58LITVDD384 pKa = 3.01GTEE387 pKa = 4.17FSLTHH392 pKa = 5.74ATSGTTAAGSIVYY405 pKa = 9.98SVTVTGSTATITLTHH420 pKa = 6.15TAGLTAAQAQALVNGIAYY438 pKa = 9.88RR439 pKa = 11.84NSSTNGMSANRR450 pKa = 11.84VVTLTEE456 pKa = 3.97ITDD459 pKa = 3.57SGIDD463 pKa = 3.64NNVTTLNLATTLQDD477 pKa = 3.32ALAPNVVSVANNVSSLVKK495 pKa = 10.37IGTGDD500 pKa = 3.16VTYY503 pKa = 9.21TYY505 pKa = 10.05TFSEE509 pKa = 4.99VVTGLTSADD518 pKa = 3.29FTVTHH523 pKa = 6.02GTVASISGSGTTWTVVVTPAANVASGKK550 pKa = 10.38LVVVLNAGSVQDD562 pKa = 3.93SVGNTSLQHH571 pKa = 5.81TDD573 pKa = 2.86STVSIDD579 pKa = 3.27TVAPVVSINTIANNDD594 pKa = 3.08IVNAAKK600 pKa = 10.24KK601 pKa = 9.63AAGVTVSGTSDD612 pKa = 3.5AEE614 pKa = 4.2VGQDD618 pKa = 4.21VSVAWGGISKK628 pKa = 8.83TGKK631 pKa = 10.0VLGDD635 pKa = 3.66GTWQVTFAAVEE646 pKa = 4.28VPGDD650 pKa = 3.8VASSTMTANVSDD662 pKa = 3.95QAGNAAVAATRR673 pKa = 11.84SVRR676 pKa = 11.84IDD678 pKa = 3.16TTAPTISITPPLTNGATVDD697 pKa = 3.63NDD699 pKa = 3.44ADD701 pKa = 4.67AILNKK706 pKa = 10.52AEE708 pKa = 4.52LDD710 pKa = 3.42AVLPNGYY717 pKa = 9.24LAVIGVTNAEE727 pKa = 4.07AGQTVTAFFNSKK739 pKa = 10.07SYY741 pKa = 9.21TATVIAGTPNNTWAVQVPKK760 pKa = 10.85ADD762 pKa = 4.22LLALAHH768 pKa = 6.33GNSYY772 pKa = 10.65SISATVSDD780 pKa = 3.87AAGNPVATPASSTLEE795 pKa = 4.14VKK797 pKa = 10.37LAPPDD802 pKa = 3.58VPTVVQLNTNDD813 pKa = 3.74LTPVITGLAQKK824 pKa = 10.1EE825 pKa = 4.19DD826 pKa = 3.85PADD829 pKa = 3.65VNSYY833 pKa = 10.26IALEE837 pKa = 4.45AGDD840 pKa = 5.19AITVTVGGQTYY851 pKa = 8.36TLTLGANAGASTPAGLTYY869 pKa = 10.65SGSTWSLAVPTALTQGQYY887 pKa = 10.91NVGVSVNAVGYY898 pKa = 7.42STPKK902 pKa = 9.65TDD904 pKa = 2.9ISSSEE909 pKa = 3.98LVIKK913 pKa = 9.92TDD915 pKa = 3.79APAITINTIAGDD927 pKa = 3.7NVINIAEE934 pKa = 4.37HH935 pKa = 6.28GGAITLSGTATDD947 pKa = 4.91NIPGVTPAVNTAAGKK962 pKa = 9.81NLSLTLAGVTYY973 pKa = 10.96SDD975 pKa = 4.5IAIQSDD981 pKa = 4.34GTWHH985 pKa = 6.31VDD987 pKa = 3.29VPAASVALLSAASYY1001 pKa = 10.26SAVVNYY1007 pKa = 9.49TGVYY1011 pKa = 10.14GNTGSQSQSLALDD1024 pKa = 3.82LVKK1027 pKa = 10.12PATPDD1032 pKa = 3.11AVLTTDD1038 pKa = 3.87TGVTDD1043 pKa = 4.13AAHH1046 pKa = 5.58VTQSAAITAPTNTEE1060 pKa = 3.32ADD1062 pKa = 2.95ATLQYY1067 pKa = 10.65RR1068 pKa = 11.84ITKK1071 pKa = 10.21SGEE1074 pKa = 3.95TEE1076 pKa = 4.3GAWGSYY1082 pKa = 9.2AAPTVDD1088 pKa = 2.66GAYY1091 pKa = 8.96TVEE1094 pKa = 4.15VRR1096 pKa = 11.84QTDD1099 pKa = 3.49AAGNPSDD1106 pKa = 3.88IQTLSIVLDD1115 pKa = 3.32NTAPVFGSLATANVNEE1131 pKa = 4.22NVAAGTVVYY1140 pKa = 8.39TANASDD1146 pKa = 3.65ANAGVVYY1153 pKa = 10.13SLKK1156 pKa = 10.87AGVGDD1161 pKa = 4.84AIKK1164 pKa = 9.38FTIDD1168 pKa = 2.89AATGQVKK1175 pKa = 9.74LQEE1178 pKa = 4.19VPNYY1182 pKa = 9.27EE1183 pKa = 4.08GQISYY1188 pKa = 11.08AFTVVATDD1196 pKa = 3.37KK1197 pKa = 11.25AGNATEE1203 pKa = 3.93QAVAVTVTDD1212 pKa = 3.72VVEE1215 pKa = 4.43VSAIAITNLSDD1226 pKa = 3.82SSVAEE1231 pKa = 3.85NAAYY1235 pKa = 8.96TSSTPSLSGTT1245 pKa = 3.57
MM1 pKa = 8.09VEE3 pKa = 3.96VSAIAITNLSDD14 pKa = 3.82SSVAEE19 pKa = 3.85NAAYY23 pKa = 8.96TSSTPSLSGTPIGAVTYY40 pKa = 10.46SLGGADD46 pKa = 3.71SALFTLNTGTGVVSMVARR64 pKa = 11.84NFEE67 pKa = 4.21SPDD70 pKa = 3.3DD71 pKa = 4.17AGANNVYY78 pKa = 10.61NYY80 pKa = 9.95TLTATDD86 pKa = 3.97ADD88 pKa = 4.47GNSADD93 pKa = 3.68QAVVLSVVDD102 pKa = 4.45FNDD105 pKa = 3.24NAPVFSSGATGSVPEE120 pKa = 4.24NAATTRR126 pKa = 11.84EE127 pKa = 4.34IYY129 pKa = 8.88TASTTDD135 pKa = 3.47ADD137 pKa = 3.63GTAANRR143 pKa = 11.84NVVYY147 pKa = 10.23SLKK150 pKa = 10.8AATGDD155 pKa = 3.51IALLDD160 pKa = 3.76IDD162 pKa = 4.22SATGKK167 pKa = 8.53VTLKK171 pKa = 10.91NSANYY176 pKa = 7.53EE177 pKa = 4.18AKK179 pKa = 10.12SSYY182 pKa = 10.87SFTVVATNVATSGTLSTEE200 pKa = 3.73QAVIANVTNVNDD212 pKa = 3.91APTVSAALASNASEE226 pKa = 4.33GDD228 pKa = 3.01SSYY231 pKa = 11.17TVNLLQGATDD241 pKa = 3.71ADD243 pKa = 3.78AGEE246 pKa = 4.3TATLSTQSLTYY257 pKa = 9.86QVNGGTASATLPAGVSITGNTLTVDD282 pKa = 3.61PANAVFNSLAFGQSQVIVASYY303 pKa = 10.66QVTDD307 pKa = 3.49VQGASVNQTATLTISGTNDD326 pKa = 3.1APTLSANTQTASYY339 pKa = 10.61ANDD342 pKa = 3.06ATAQTVFSGAFISAVDD358 pKa = 3.62AGQTIKK364 pKa = 10.88GLKK367 pKa = 8.54FTVSGITDD375 pKa = 3.75ASSEE379 pKa = 4.58LITVDD384 pKa = 3.01GTEE387 pKa = 4.17FSLTHH392 pKa = 5.74ATSGTTAAGSIVYY405 pKa = 9.98SVTVTGSTATITLTHH420 pKa = 6.15TAGLTAAQAQALVNGIAYY438 pKa = 9.88RR439 pKa = 11.84NSSTNGMSANRR450 pKa = 11.84VVTLTEE456 pKa = 3.97ITDD459 pKa = 3.57SGIDD463 pKa = 3.64NNVTTLNLATTLQDD477 pKa = 3.32ALAPNVVSVANNVSSLVKK495 pKa = 10.37IGTGDD500 pKa = 3.16VTYY503 pKa = 9.21TYY505 pKa = 10.05TFSEE509 pKa = 4.99VVTGLTSADD518 pKa = 3.29FTVTHH523 pKa = 6.02GTVASISGSGTTWTVVVTPAANVASGKK550 pKa = 10.38LVVVLNAGSVQDD562 pKa = 3.93SVGNTSLQHH571 pKa = 5.81TDD573 pKa = 2.86STVSIDD579 pKa = 3.27TVAPVVSINTIANNDD594 pKa = 3.08IVNAAKK600 pKa = 10.24KK601 pKa = 9.63AAGVTVSGTSDD612 pKa = 3.5AEE614 pKa = 4.2VGQDD618 pKa = 4.21VSVAWGGISKK628 pKa = 8.83TGKK631 pKa = 10.0VLGDD635 pKa = 3.66GTWQVTFAAVEE646 pKa = 4.28VPGDD650 pKa = 3.8VASSTMTANVSDD662 pKa = 3.95QAGNAAVAATRR673 pKa = 11.84SVRR676 pKa = 11.84IDD678 pKa = 3.16TTAPTISITPPLTNGATVDD697 pKa = 3.63NDD699 pKa = 3.44ADD701 pKa = 4.67AILNKK706 pKa = 10.52AEE708 pKa = 4.52LDD710 pKa = 3.42AVLPNGYY717 pKa = 9.24LAVIGVTNAEE727 pKa = 4.07AGQTVTAFFNSKK739 pKa = 10.07SYY741 pKa = 9.21TATVIAGTPNNTWAVQVPKK760 pKa = 10.85ADD762 pKa = 4.22LLALAHH768 pKa = 6.33GNSYY772 pKa = 10.65SISATVSDD780 pKa = 3.87AAGNPVATPASSTLEE795 pKa = 4.14VKK797 pKa = 10.37LAPPDD802 pKa = 3.58VPTVVQLNTNDD813 pKa = 3.74LTPVITGLAQKK824 pKa = 10.1EE825 pKa = 4.19DD826 pKa = 3.85PADD829 pKa = 3.65VNSYY833 pKa = 10.26IALEE837 pKa = 4.45AGDD840 pKa = 5.19AITVTVGGQTYY851 pKa = 8.36TLTLGANAGASTPAGLTYY869 pKa = 10.65SGSTWSLAVPTALTQGQYY887 pKa = 10.91NVGVSVNAVGYY898 pKa = 7.42STPKK902 pKa = 9.65TDD904 pKa = 2.9ISSSEE909 pKa = 3.98LVIKK913 pKa = 9.92TDD915 pKa = 3.79APAITINTIAGDD927 pKa = 3.7NVINIAEE934 pKa = 4.37HH935 pKa = 6.28GGAITLSGTATDD947 pKa = 4.91NIPGVTPAVNTAAGKK962 pKa = 9.81NLSLTLAGVTYY973 pKa = 10.96SDD975 pKa = 4.5IAIQSDD981 pKa = 4.34GTWHH985 pKa = 6.31VDD987 pKa = 3.29VPAASVALLSAASYY1001 pKa = 10.26SAVVNYY1007 pKa = 9.49TGVYY1011 pKa = 10.14GNTGSQSQSLALDD1024 pKa = 3.82LVKK1027 pKa = 10.12PATPDD1032 pKa = 3.11AVLTTDD1038 pKa = 3.87TGVTDD1043 pKa = 4.13AAHH1046 pKa = 5.58VTQSAAITAPTNTEE1060 pKa = 3.32ADD1062 pKa = 2.95ATLQYY1067 pKa = 10.65RR1068 pKa = 11.84ITKK1071 pKa = 10.21SGEE1074 pKa = 3.95TEE1076 pKa = 4.3GAWGSYY1082 pKa = 9.2AAPTVDD1088 pKa = 2.66GAYY1091 pKa = 8.96TVEE1094 pKa = 4.15VRR1096 pKa = 11.84QTDD1099 pKa = 3.49AAGNPSDD1106 pKa = 3.88IQTLSIVLDD1115 pKa = 3.32NTAPVFGSLATANVNEE1131 pKa = 4.22NVAAGTVVYY1140 pKa = 8.39TANASDD1146 pKa = 3.65ANAGVVYY1153 pKa = 10.13SLKK1156 pKa = 10.87AGVGDD1161 pKa = 4.84AIKK1164 pKa = 9.38FTIDD1168 pKa = 2.89AATGQVKK1175 pKa = 9.74LQEE1178 pKa = 4.19VPNYY1182 pKa = 9.27EE1183 pKa = 4.08GQISYY1188 pKa = 11.08AFTVVATDD1196 pKa = 3.37KK1197 pKa = 11.25AGNATEE1203 pKa = 3.93QAVAVTVTDD1212 pKa = 3.72VVEE1215 pKa = 4.43VSAIAITNLSDD1226 pKa = 3.82SSVAEE1231 pKa = 3.85NAAYY1235 pKa = 8.96TSSTPSLSGTT1245 pKa = 3.57
Molecular weight: 124.59 kDa
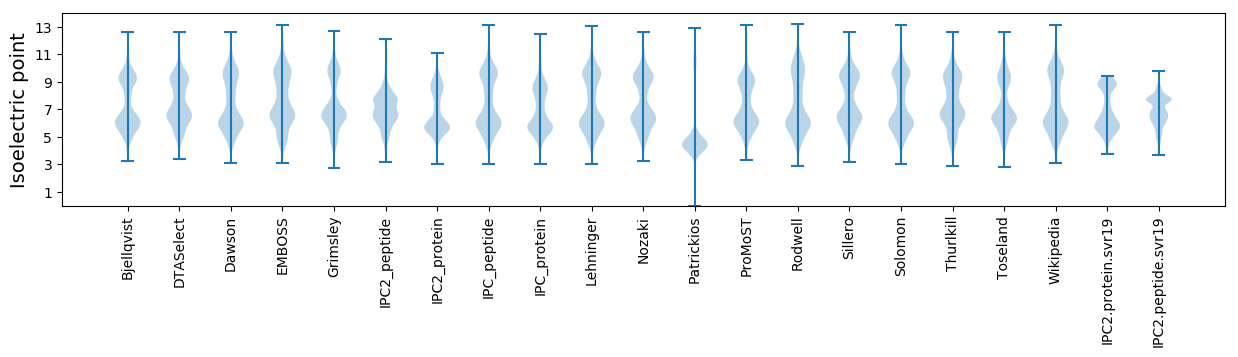
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A315EL79|A0A315EL79_9BURK Aldehyde-activating protein OS=Limnohabitans sp. 2KL-17 OX=1100704 GN=B9Z45_07890 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1042381 |
31 |
4604 |
327.4 |
35.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.705 ± 0.064 | 0.972 ± 0.016 |
5.108 ± 0.033 | 5.117 ± 0.041 |
3.56 ± 0.029 | 8.084 ± 0.052 |
2.364 ± 0.025 | 4.582 ± 0.039 |
4.053 ± 0.048 | 10.7 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.796 ± 0.026 | 2.999 ± 0.035 |
4.95 ± 0.036 | 4.653 ± 0.032 |
5.769 ± 0.048 | 5.767 ± 0.045 |
5.48 ± 0.051 | 7.613 ± 0.032 |
1.563 ± 0.021 | 2.165 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
