
Lacimicrobium alkaliphilum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Lacimicrobium
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
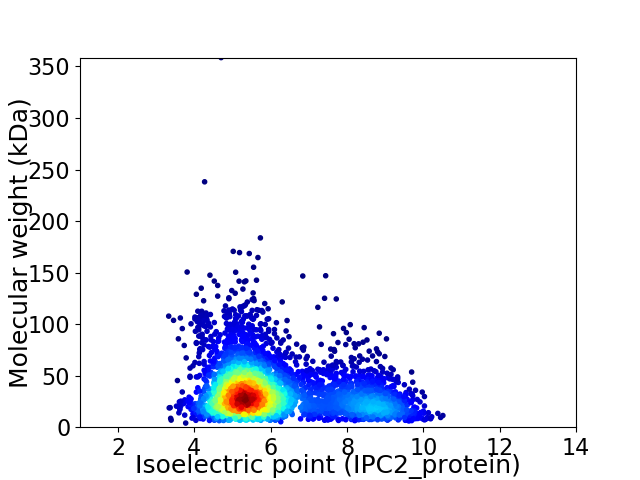
Virtual 2D-PAGE plot for 3754 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
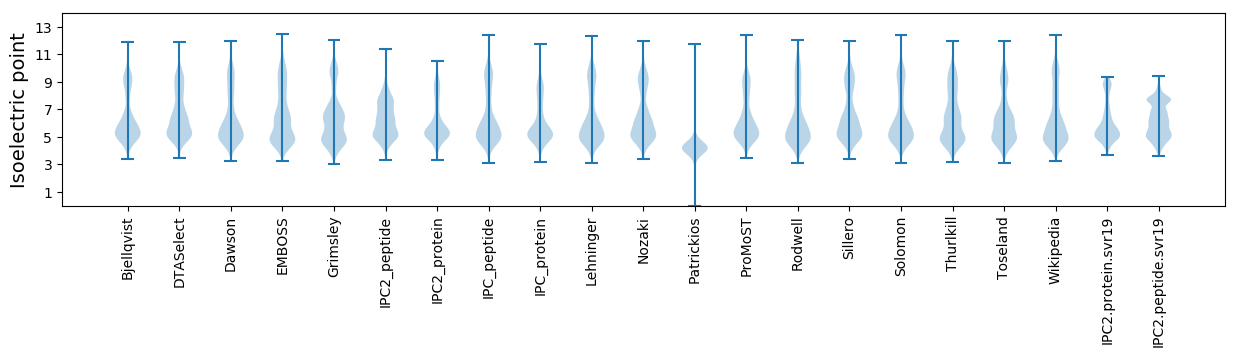
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U3ALQ6|A0A0U3ALQ6_9ALTE Uncharacterized protein OS=Lacimicrobium alkaliphilum OX=1526571 GN=AT746_11950 PE=4 SV=1
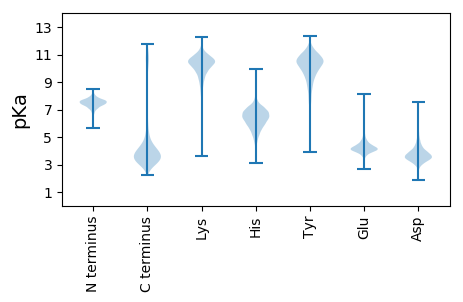
MM1 pKa = 7.07QVTAAPADD9 pKa = 3.59SLIISGVVDD18 pKa = 4.55GPLPGGLPKK27 pKa = 10.48AVEE30 pKa = 4.3VYY32 pKa = 9.45VAKK35 pKa = 10.83DD36 pKa = 2.86IDD38 pKa = 5.23DD39 pKa = 3.96ISQCGLGSANNGGGTDD55 pKa = 3.7GEE57 pKa = 4.61EE58 pKa = 3.97FTFPSGAVSAGQYY71 pKa = 10.36LYY73 pKa = 10.9VASEE77 pKa = 4.33SAGFTDD83 pKa = 5.92FFGFSPNYY91 pKa = 8.66TSSAANINGDD101 pKa = 3.61DD102 pKa = 4.74AIEE105 pKa = 4.18LFCDD109 pKa = 3.97GAVVDD114 pKa = 4.45TFGEE118 pKa = 4.51TEE120 pKa = 3.87VDD122 pKa = 3.12GSGQVWEE129 pKa = 4.79YY130 pKa = 11.14LDD132 pKa = 3.19GWAYY136 pKa = 10.13RR137 pKa = 11.84VAEE140 pKa = 4.31TGPDD144 pKa = 2.97AGNFVQANWQFSGPNALDD162 pKa = 3.75NEE164 pKa = 4.58SSNASAAVPFPLATFAGDD182 pKa = 3.84GGDD185 pKa = 3.81TPVDD189 pKa = 4.77PIDD192 pKa = 5.29PDD194 pKa = 5.17PIDD197 pKa = 4.93PDD199 pKa = 3.61PVEE202 pKa = 4.45PGEE205 pKa = 4.39GGEE208 pKa = 4.08GLIFSEE214 pKa = 4.6YY215 pKa = 10.95VEE217 pKa = 4.23GSSFNKK223 pKa = 10.25ALEE226 pKa = 4.27LANTTDD232 pKa = 3.07STLDD236 pKa = 3.3LGGYY240 pKa = 8.34QVLLYY245 pKa = 10.95SNSNTTAGTTLDD257 pKa = 3.88LSGSLAAGEE266 pKa = 4.3VLVIANSQAGDD277 pKa = 4.67AIQSVADD284 pKa = 3.46IVSGVTNFNGNDD296 pKa = 3.65ALEE299 pKa = 4.08LRR301 pKa = 11.84YY302 pKa = 9.99NDD304 pKa = 4.32EE305 pKa = 4.51LVDD308 pKa = 4.41SIGQVGSGQYY318 pKa = 10.03FGSNVTLVRR327 pKa = 11.84KK328 pKa = 9.92ADD330 pKa = 3.35ITAGDD335 pKa = 4.1TDD337 pKa = 3.91SSDD340 pKa = 3.42TFVAADD346 pKa = 3.27QWQEE350 pKa = 3.81FASDD354 pKa = 3.16TFTYY358 pKa = 10.33LGSHH362 pKa = 6.39NGDD365 pKa = 4.09DD366 pKa = 4.77GGDD369 pKa = 3.55DD370 pKa = 3.79TPVVEE375 pKa = 5.86LGQCGDD381 pKa = 3.7PATLISQVQGSGDD394 pKa = 3.47VSPVVSEE401 pKa = 3.77SHH403 pKa = 6.85IIEE406 pKa = 4.24AVVTGTFTNLQGYY419 pKa = 7.49FVQEE423 pKa = 4.43EE424 pKa = 4.26VADD427 pKa = 3.94WDD429 pKa = 3.94NDD431 pKa = 3.16ASTSEE436 pKa = 4.19GVFVYY441 pKa = 10.36TNSDD445 pKa = 3.14QNLPQPGTLVRR456 pKa = 11.84VQGEE460 pKa = 3.99VSEE463 pKa = 5.47FFDD466 pKa = 3.8KK467 pKa = 10.66TQLTATASALDD478 pKa = 4.15CGQADD483 pKa = 3.99TATVSLSLPFADD495 pKa = 4.0AQQTEE500 pKa = 4.61TLEE503 pKa = 4.2GMLVSFDD510 pKa = 3.84HH511 pKa = 6.67SLTVTDD517 pKa = 4.28NYY519 pKa = 11.43NLGRR523 pKa = 11.84YY524 pKa = 9.76GEE526 pKa = 4.41VTLSNGRR533 pKa = 11.84TYY535 pKa = 10.82IPTNLHH541 pKa = 6.25LPGSPEE547 pKa = 3.74AQALAEE553 pKa = 4.02QNALNRR559 pKa = 11.84ILLDD563 pKa = 4.75DD564 pKa = 5.7GINGQNPATVIYY576 pKa = 7.3PTGGLSAGNTLRR588 pKa = 11.84TGDD591 pKa = 4.03EE592 pKa = 4.17VSALTGVLDD601 pKa = 3.9YY602 pKa = 11.71GFGEE606 pKa = 4.17YY607 pKa = 10.07RR608 pKa = 11.84IIPVEE613 pKa = 4.04APTFVASNPRR623 pKa = 11.84TPAPEE628 pKa = 3.71IAEE631 pKa = 4.46GNLKK635 pKa = 10.11VASLNVLNFFNGNGLGGGFPTPRR658 pKa = 11.84GADD661 pKa = 3.04NVEE664 pKa = 4.17EE665 pKa = 4.25YY666 pKa = 10.44QRR668 pKa = 11.84QLDD671 pKa = 4.0KK672 pKa = 11.21LVSAILAMDD681 pKa = 4.56ADD683 pKa = 3.97IVGLMEE689 pKa = 5.03IEE691 pKa = 4.07NDD693 pKa = 4.49GYY695 pKa = 11.26DD696 pKa = 3.51QNSAIAQLTDD706 pKa = 3.47ALNAQAGDD714 pKa = 3.4AVYY717 pKa = 10.45QYY719 pKa = 11.4VNAGGPLGTDD729 pKa = 4.24AIAVGLLYY737 pKa = 10.49KK738 pKa = 10.73ANTVTPVGDD747 pKa = 3.56VRR749 pKa = 11.84IHH751 pKa = 7.01LDD753 pKa = 3.26EE754 pKa = 4.71VYY756 pKa = 10.75NRR758 pKa = 11.84PPLAQRR764 pKa = 11.84FSNTEE769 pKa = 3.68GGEE772 pKa = 3.74LSVIVNHH779 pKa = 6.41FKK781 pKa = 11.2SKK783 pKa = 10.38GGCGSAEE790 pKa = 4.92GLDD793 pKa = 3.62QDD795 pKa = 4.66QGDD798 pKa = 4.05GQACFNATRR807 pKa = 11.84VAQAQALLDD816 pKa = 4.26WIATDD821 pKa = 3.63EE822 pKa = 4.1QLSGEE827 pKa = 4.26EE828 pKa = 3.77NHH830 pKa = 7.1LVIGDD835 pKa = 4.0LNAYY839 pKa = 9.88AKK841 pKa = 9.82EE842 pKa = 3.98DD843 pKa = 4.51PIRR846 pKa = 11.84EE847 pKa = 4.17FTDD850 pKa = 2.97NGYY853 pKa = 8.57TNLIEE858 pKa = 5.09AVAGPQAYY866 pKa = 9.68SYY868 pKa = 11.32VFAGEE873 pKa = 4.03AGYY876 pKa = 10.58LDD878 pKa = 3.93HH879 pKa = 7.55AIASDD884 pKa = 3.42ALAAQVVDD892 pKa = 3.54VTEE895 pKa = 3.66WHH897 pKa = 6.81INADD901 pKa = 3.6EE902 pKa = 4.31PRR904 pKa = 11.84ILDD907 pKa = 3.51YY908 pKa = 10.31NTEE911 pKa = 4.27FKK913 pKa = 10.89SQQQVLDD920 pKa = 4.98FYY922 pKa = 11.79APDD925 pKa = 3.44SYY927 pKa = 10.81RR928 pKa = 11.84ASDD931 pKa = 4.01HH932 pKa = 7.18DD933 pKa = 4.17PVIVTLDD940 pKa = 3.43LQAAEE945 pKa = 4.37LQGDD949 pKa = 4.06FDD951 pKa = 5.49GDD953 pKa = 3.81GDD955 pKa = 4.21VDD957 pKa = 4.19SSDD960 pKa = 3.35MKK962 pKa = 11.37GLMRR966 pKa = 11.84AILTRR971 pKa = 11.84QSIDD975 pKa = 3.01MAFDD979 pKa = 4.22LNGDD983 pKa = 3.75GVINTRR989 pKa = 11.84DD990 pKa = 3.59LQILFTLCTRR1000 pKa = 11.84ANCATEE1006 pKa = 3.7
MM1 pKa = 7.07QVTAAPADD9 pKa = 3.59SLIISGVVDD18 pKa = 4.55GPLPGGLPKK27 pKa = 10.48AVEE30 pKa = 4.3VYY32 pKa = 9.45VAKK35 pKa = 10.83DD36 pKa = 2.86IDD38 pKa = 5.23DD39 pKa = 3.96ISQCGLGSANNGGGTDD55 pKa = 3.7GEE57 pKa = 4.61EE58 pKa = 3.97FTFPSGAVSAGQYY71 pKa = 10.36LYY73 pKa = 10.9VASEE77 pKa = 4.33SAGFTDD83 pKa = 5.92FFGFSPNYY91 pKa = 8.66TSSAANINGDD101 pKa = 3.61DD102 pKa = 4.74AIEE105 pKa = 4.18LFCDD109 pKa = 3.97GAVVDD114 pKa = 4.45TFGEE118 pKa = 4.51TEE120 pKa = 3.87VDD122 pKa = 3.12GSGQVWEE129 pKa = 4.79YY130 pKa = 11.14LDD132 pKa = 3.19GWAYY136 pKa = 10.13RR137 pKa = 11.84VAEE140 pKa = 4.31TGPDD144 pKa = 2.97AGNFVQANWQFSGPNALDD162 pKa = 3.75NEE164 pKa = 4.58SSNASAAVPFPLATFAGDD182 pKa = 3.84GGDD185 pKa = 3.81TPVDD189 pKa = 4.77PIDD192 pKa = 5.29PDD194 pKa = 5.17PIDD197 pKa = 4.93PDD199 pKa = 3.61PVEE202 pKa = 4.45PGEE205 pKa = 4.39GGEE208 pKa = 4.08GLIFSEE214 pKa = 4.6YY215 pKa = 10.95VEE217 pKa = 4.23GSSFNKK223 pKa = 10.25ALEE226 pKa = 4.27LANTTDD232 pKa = 3.07STLDD236 pKa = 3.3LGGYY240 pKa = 8.34QVLLYY245 pKa = 10.95SNSNTTAGTTLDD257 pKa = 3.88LSGSLAAGEE266 pKa = 4.3VLVIANSQAGDD277 pKa = 4.67AIQSVADD284 pKa = 3.46IVSGVTNFNGNDD296 pKa = 3.65ALEE299 pKa = 4.08LRR301 pKa = 11.84YY302 pKa = 9.99NDD304 pKa = 4.32EE305 pKa = 4.51LVDD308 pKa = 4.41SIGQVGSGQYY318 pKa = 10.03FGSNVTLVRR327 pKa = 11.84KK328 pKa = 9.92ADD330 pKa = 3.35ITAGDD335 pKa = 4.1TDD337 pKa = 3.91SSDD340 pKa = 3.42TFVAADD346 pKa = 3.27QWQEE350 pKa = 3.81FASDD354 pKa = 3.16TFTYY358 pKa = 10.33LGSHH362 pKa = 6.39NGDD365 pKa = 4.09DD366 pKa = 4.77GGDD369 pKa = 3.55DD370 pKa = 3.79TPVVEE375 pKa = 5.86LGQCGDD381 pKa = 3.7PATLISQVQGSGDD394 pKa = 3.47VSPVVSEE401 pKa = 3.77SHH403 pKa = 6.85IIEE406 pKa = 4.24AVVTGTFTNLQGYY419 pKa = 7.49FVQEE423 pKa = 4.43EE424 pKa = 4.26VADD427 pKa = 3.94WDD429 pKa = 3.94NDD431 pKa = 3.16ASTSEE436 pKa = 4.19GVFVYY441 pKa = 10.36TNSDD445 pKa = 3.14QNLPQPGTLVRR456 pKa = 11.84VQGEE460 pKa = 3.99VSEE463 pKa = 5.47FFDD466 pKa = 3.8KK467 pKa = 10.66TQLTATASALDD478 pKa = 4.15CGQADD483 pKa = 3.99TATVSLSLPFADD495 pKa = 4.0AQQTEE500 pKa = 4.61TLEE503 pKa = 4.2GMLVSFDD510 pKa = 3.84HH511 pKa = 6.67SLTVTDD517 pKa = 4.28NYY519 pKa = 11.43NLGRR523 pKa = 11.84YY524 pKa = 9.76GEE526 pKa = 4.41VTLSNGRR533 pKa = 11.84TYY535 pKa = 10.82IPTNLHH541 pKa = 6.25LPGSPEE547 pKa = 3.74AQALAEE553 pKa = 4.02QNALNRR559 pKa = 11.84ILLDD563 pKa = 4.75DD564 pKa = 5.7GINGQNPATVIYY576 pKa = 7.3PTGGLSAGNTLRR588 pKa = 11.84TGDD591 pKa = 4.03EE592 pKa = 4.17VSALTGVLDD601 pKa = 3.9YY602 pKa = 11.71GFGEE606 pKa = 4.17YY607 pKa = 10.07RR608 pKa = 11.84IIPVEE613 pKa = 4.04APTFVASNPRR623 pKa = 11.84TPAPEE628 pKa = 3.71IAEE631 pKa = 4.46GNLKK635 pKa = 10.11VASLNVLNFFNGNGLGGGFPTPRR658 pKa = 11.84GADD661 pKa = 3.04NVEE664 pKa = 4.17EE665 pKa = 4.25YY666 pKa = 10.44QRR668 pKa = 11.84QLDD671 pKa = 4.0KK672 pKa = 11.21LVSAILAMDD681 pKa = 4.56ADD683 pKa = 3.97IVGLMEE689 pKa = 5.03IEE691 pKa = 4.07NDD693 pKa = 4.49GYY695 pKa = 11.26DD696 pKa = 3.51QNSAIAQLTDD706 pKa = 3.47ALNAQAGDD714 pKa = 3.4AVYY717 pKa = 10.45QYY719 pKa = 11.4VNAGGPLGTDD729 pKa = 4.24AIAVGLLYY737 pKa = 10.49KK738 pKa = 10.73ANTVTPVGDD747 pKa = 3.56VRR749 pKa = 11.84IHH751 pKa = 7.01LDD753 pKa = 3.26EE754 pKa = 4.71VYY756 pKa = 10.75NRR758 pKa = 11.84PPLAQRR764 pKa = 11.84FSNTEE769 pKa = 3.68GGEE772 pKa = 3.74LSVIVNHH779 pKa = 6.41FKK781 pKa = 11.2SKK783 pKa = 10.38GGCGSAEE790 pKa = 4.92GLDD793 pKa = 3.62QDD795 pKa = 4.66QGDD798 pKa = 4.05GQACFNATRR807 pKa = 11.84VAQAQALLDD816 pKa = 4.26WIATDD821 pKa = 3.63EE822 pKa = 4.1QLSGEE827 pKa = 4.26EE828 pKa = 3.77NHH830 pKa = 7.1LVIGDD835 pKa = 4.0LNAYY839 pKa = 9.88AKK841 pKa = 9.82EE842 pKa = 3.98DD843 pKa = 4.51PIRR846 pKa = 11.84EE847 pKa = 4.17FTDD850 pKa = 2.97NGYY853 pKa = 8.57TNLIEE858 pKa = 5.09AVAGPQAYY866 pKa = 9.68SYY868 pKa = 11.32VFAGEE873 pKa = 4.03AGYY876 pKa = 10.58LDD878 pKa = 3.93HH879 pKa = 7.55AIASDD884 pKa = 3.42ALAAQVVDD892 pKa = 3.54VTEE895 pKa = 3.66WHH897 pKa = 6.81INADD901 pKa = 3.6EE902 pKa = 4.31PRR904 pKa = 11.84ILDD907 pKa = 3.51YY908 pKa = 10.31NTEE911 pKa = 4.27FKK913 pKa = 10.89SQQQVLDD920 pKa = 4.98FYY922 pKa = 11.79APDD925 pKa = 3.44SYY927 pKa = 10.81RR928 pKa = 11.84ASDD931 pKa = 4.01HH932 pKa = 7.18DD933 pKa = 4.17PVIVTLDD940 pKa = 3.43LQAAEE945 pKa = 4.37LQGDD949 pKa = 4.06FDD951 pKa = 5.49GDD953 pKa = 3.81GDD955 pKa = 4.21VDD957 pKa = 4.19SSDD960 pKa = 3.35MKK962 pKa = 11.37GLMRR966 pKa = 11.84AILTRR971 pKa = 11.84QSIDD975 pKa = 3.01MAFDD979 pKa = 4.22LNGDD983 pKa = 3.75GVINTRR989 pKa = 11.84DD990 pKa = 3.59LQILFTLCTRR1000 pKa = 11.84ANCATEE1006 pKa = 3.7
Molecular weight: 105.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U3B2A8|A0A0U3B2A8_9ALTE HTH araC/xylS-type domain-containing protein OS=Lacimicrobium alkaliphilum OX=1526571 GN=AT746_14710 PE=4 SV=1
MM1 pKa = 7.15QFIAKK6 pKa = 8.73IQSARR11 pKa = 11.84SFRR14 pKa = 11.84NCNTALLRR22 pKa = 11.84ALCDD26 pKa = 3.45SAVRR30 pKa = 11.84NAFFSGLSLSYY41 pKa = 11.12ALRR44 pKa = 11.84AAIKK48 pKa = 10.36RR49 pKa = 11.84LALKK53 pKa = 9.89IAPGDD58 pKa = 3.65FFVSFVVKK66 pKa = 9.99KK67 pKa = 9.46HH68 pKa = 4.53WANILGSPINWTLHH82 pKa = 7.08DD83 pKa = 4.01IALLPALCAPAPLRR97 pKa = 11.84EE98 pKa = 4.29PKK100 pKa = 10.7LSFSDD105 pKa = 3.49LAEE108 pKa = 4.05SLRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AGKK116 pKa = 9.96
MM1 pKa = 7.15QFIAKK6 pKa = 8.73IQSARR11 pKa = 11.84SFRR14 pKa = 11.84NCNTALLRR22 pKa = 11.84ALCDD26 pKa = 3.45SAVRR30 pKa = 11.84NAFFSGLSLSYY41 pKa = 11.12ALRR44 pKa = 11.84AAIKK48 pKa = 10.36RR49 pKa = 11.84LALKK53 pKa = 9.89IAPGDD58 pKa = 3.65FFVSFVVKK66 pKa = 9.99KK67 pKa = 9.46HH68 pKa = 4.53WANILGSPINWTLHH82 pKa = 7.08DD83 pKa = 4.01IALLPALCAPAPLRR97 pKa = 11.84EE98 pKa = 4.29PKK100 pKa = 10.7LSFSDD105 pKa = 3.49LAEE108 pKa = 4.05SLRR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AGKK116 pKa = 9.96
Molecular weight: 12.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1254132 |
34 |
3323 |
334.1 |
37.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.018 ± 0.043 | 1.011 ± 0.014 |
5.724 ± 0.034 | 6.111 ± 0.039 |
3.988 ± 0.026 | 7.138 ± 0.039 |
2.283 ± 0.023 | 5.662 ± 0.028 |
4.508 ± 0.036 | 11.002 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.562 ± 0.018 | 3.805 ± 0.023 |
4.202 ± 0.025 | 5.452 ± 0.036 |
5.221 ± 0.025 | 6.414 ± 0.031 |
4.867 ± 0.027 | 6.556 ± 0.032 |
1.389 ± 0.018 | 3.087 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
