
Sclerophthora macrospora virus A
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA positive-strand viruses
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins
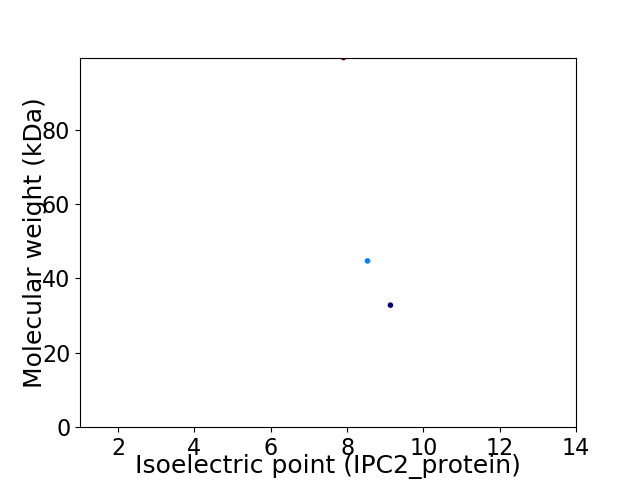
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8BET0|Q8BET0_9VIRU RNA replicase OS=Sclerophthora macrospora virus A OX=191289 PE=3 SV=1
MM1 pKa = 7.29PWKK4 pKa = 9.94PGKK7 pKa = 9.97CHH9 pKa = 7.26LLAAGLAGVCCIAYY23 pKa = 7.41TKK25 pKa = 10.48RR26 pKa = 11.84GSLRR30 pKa = 11.84DD31 pKa = 3.46AASALCEE38 pKa = 4.04WLADD42 pKa = 3.89EE43 pKa = 4.61PAIVSDD49 pKa = 3.93LSRR52 pKa = 11.84DD53 pKa = 3.6AFTTTCVDD61 pKa = 3.37PVVAISGHH69 pKa = 4.23THH71 pKa = 6.69ASAASLRR78 pKa = 11.84TAATRR83 pKa = 11.84YY84 pKa = 8.07AQNVAQYY91 pKa = 9.57CGTEE95 pKa = 4.27VYY97 pKa = 10.26VVGMSRR103 pKa = 11.84SDD105 pKa = 3.13QRR107 pKa = 11.84KK108 pKa = 9.02DD109 pKa = 3.36MKK111 pKa = 10.79GSRR114 pKa = 11.84QWYY117 pKa = 4.31WTKK120 pKa = 11.1DD121 pKa = 3.23VNAANRR127 pKa = 11.84NDD129 pKa = 3.96RR130 pKa = 11.84PGHH133 pKa = 5.58RR134 pKa = 11.84DD135 pKa = 2.71IRR137 pKa = 11.84FLCDD141 pKa = 2.45VDD143 pKa = 5.16YY144 pKa = 11.08YY145 pKa = 11.83VDD147 pKa = 3.79MPAMLVAEE155 pKa = 5.23AKK157 pKa = 10.01PVLLYY162 pKa = 10.21TVVPEE167 pKa = 4.32EE168 pKa = 4.34ATSSANDD175 pKa = 3.02EE176 pKa = 4.23SAFYY180 pKa = 10.78FEE182 pKa = 5.82ADD184 pKa = 3.08GSLRR188 pKa = 11.84TMVAGGGGYY197 pKa = 7.35SHH199 pKa = 7.39CIWDD203 pKa = 3.7YY204 pKa = 11.32AADD207 pKa = 3.63SFLVVKK213 pKa = 9.17KK214 pKa = 10.28ICGVPIRR221 pKa = 11.84AVAYY225 pKa = 9.43AVEE228 pKa = 4.39RR229 pKa = 11.84KK230 pKa = 8.59QVGKK234 pKa = 9.65HH235 pKa = 4.19RR236 pKa = 11.84QMVLLAPVRR245 pKa = 11.84AFSGLAAILAYY256 pKa = 10.69LLLEE260 pKa = 4.32TKK262 pKa = 8.77EE263 pKa = 3.97LRR265 pKa = 11.84RR266 pKa = 11.84FNPIVEE272 pKa = 4.3AGGEE276 pKa = 4.03KK277 pKa = 9.88FVRR280 pKa = 11.84FNVMSSDD287 pKa = 3.32GALMVTTARR296 pKa = 11.84PGSSLCATVSCADD309 pKa = 4.13DD310 pKa = 4.23DD311 pKa = 5.66AIATVARR318 pKa = 11.84LGSTNLMLPTTASWVKK334 pKa = 10.33DD335 pKa = 3.94RR336 pKa = 11.84PAAAVLTDD344 pKa = 3.36YY345 pKa = 10.84HH346 pKa = 6.78RR347 pKa = 11.84KK348 pKa = 9.29CGKK351 pKa = 8.57KK352 pKa = 10.21AKK354 pKa = 9.67FVVYY358 pKa = 9.47PVEE361 pKa = 4.05QGVRR365 pKa = 11.84AYY367 pKa = 10.17QYY369 pKa = 11.21KK370 pKa = 10.21PDD372 pKa = 4.42EE373 pKa = 4.71FDD375 pKa = 2.84QAARR379 pKa = 11.84PKK381 pKa = 9.8LQAFMSPIVHH391 pKa = 6.5GAFAPVLNRR400 pKa = 11.84AGEE403 pKa = 4.13EE404 pKa = 3.93RR405 pKa = 11.84CVEE408 pKa = 3.86GRR410 pKa = 11.84INSLRR415 pKa = 11.84KK416 pKa = 9.65PEE418 pKa = 3.88PHH420 pKa = 6.95PSNFRR425 pKa = 11.84DD426 pKa = 3.28RR427 pKa = 11.84CIDD430 pKa = 3.4EE431 pKa = 4.59FADD434 pKa = 3.92LVVSGMLLEE443 pKa = 4.38PVCFEE448 pKa = 4.32VVNSKK453 pKa = 7.44QTRR456 pKa = 11.84STQKK460 pKa = 10.64LSIAKK465 pKa = 9.8AVLTGKK471 pKa = 9.85FRR473 pKa = 11.84QAVLKK478 pKa = 10.86CFGKK482 pKa = 10.66AEE484 pKa = 4.58AYY486 pKa = 9.68PDD488 pKa = 3.53VKK490 pKa = 10.81DD491 pKa = 3.85PRR493 pKa = 11.84NISQYY498 pKa = 11.4NDD500 pKa = 3.26ADD502 pKa = 4.14KK503 pKa = 11.63LDD505 pKa = 3.67MATFALALSEE515 pKa = 4.67HH516 pKa = 6.08MKK518 pKa = 9.62QFKK521 pKa = 9.69WYY523 pKa = 10.11GPGKK527 pKa = 8.81TPLEE531 pKa = 3.85IANRR535 pKa = 11.84VVEE538 pKa = 4.43ICQAADD544 pKa = 3.41FVNISDD550 pKa = 3.79YY551 pKa = 11.35HH552 pKa = 8.03RR553 pKa = 11.84MDD555 pKa = 3.0GTITYY560 pKa = 7.32TLRR563 pKa = 11.84RR564 pKa = 11.84VEE566 pKa = 4.33RR567 pKa = 11.84AVCMKK572 pKa = 10.8AFANHH577 pKa = 5.83SACLNEE583 pKa = 3.97LLKK586 pKa = 11.17RR587 pKa = 11.84NVDD590 pKa = 3.34NVGYY594 pKa = 9.55LPHH597 pKa = 6.74GTTFNQGSSHH607 pKa = 6.79GSGCSATSLFQTLRR621 pKa = 11.84AAFNAYY627 pKa = 10.22LGFRR631 pKa = 11.84HH632 pKa = 5.86TRR634 pKa = 11.84NQAGRR639 pKa = 11.84MFSPKK644 pKa = 9.86EE645 pKa = 3.72AFASLGIHH653 pKa = 6.81LGDD656 pKa = 5.87DD657 pKa = 3.98GLDD660 pKa = 3.49GNLPIGSHH668 pKa = 4.91QWASKK673 pKa = 7.08QTGLILEE680 pKa = 4.38AALVYY685 pKa = 10.21RR686 pKa = 11.84GYY688 pKa = 10.82RR689 pKa = 11.84GVNFLARR696 pKa = 11.84YY697 pKa = 7.47YY698 pKa = 10.9SPEE701 pKa = 3.69VWQGCPDD708 pKa = 3.63SMCDD712 pKa = 4.07FKK714 pKa = 11.42RR715 pKa = 11.84QISKK719 pKa = 9.63FHH721 pKa = 5.25TTVRR725 pKa = 11.84LPSNVTPEE733 pKa = 3.87QKK735 pKa = 10.43LVEE738 pKa = 4.08KK739 pKa = 10.74SMSYY743 pKa = 10.74VATDD747 pKa = 3.02GNTPVIGQLCKK758 pKa = 10.16RR759 pKa = 11.84VLLLSSLRR767 pKa = 11.84PRR769 pKa = 11.84TLFGLGNWWSQFDD782 pKa = 5.25DD783 pKa = 4.0SVQFPNNNVGGWMDD797 pKa = 3.73VEE799 pKa = 4.73IDD801 pKa = 3.55HH802 pKa = 6.44QFPEE806 pKa = 4.27FDD808 pKa = 4.17RR809 pKa = 11.84NLFDD813 pKa = 3.38KK814 pKa = 10.63WLDD817 pKa = 3.71SVQKK821 pKa = 11.19AEE823 pKa = 4.9DD824 pKa = 3.93LLSAPLCAEE833 pKa = 4.92PKK835 pKa = 10.07PPTPGCVAVVVDD847 pKa = 4.85EE848 pKa = 4.94EE849 pKa = 4.71IIPPRR854 pKa = 11.84QDD856 pKa = 2.98VEE858 pKa = 4.28PARR861 pKa = 11.84SSSPPRR867 pKa = 11.84RR868 pKa = 11.84TPLRR872 pKa = 11.84PTRR875 pKa = 11.84YY876 pKa = 8.22KK877 pKa = 9.89VTVARR882 pKa = 11.84EE883 pKa = 3.81KK884 pKa = 10.51ATLLNPPKK892 pKa = 9.87PRR894 pKa = 11.84RR895 pKa = 11.84ATAA898 pKa = 3.35
MM1 pKa = 7.29PWKK4 pKa = 9.94PGKK7 pKa = 9.97CHH9 pKa = 7.26LLAAGLAGVCCIAYY23 pKa = 7.41TKK25 pKa = 10.48RR26 pKa = 11.84GSLRR30 pKa = 11.84DD31 pKa = 3.46AASALCEE38 pKa = 4.04WLADD42 pKa = 3.89EE43 pKa = 4.61PAIVSDD49 pKa = 3.93LSRR52 pKa = 11.84DD53 pKa = 3.6AFTTTCVDD61 pKa = 3.37PVVAISGHH69 pKa = 4.23THH71 pKa = 6.69ASAASLRR78 pKa = 11.84TAATRR83 pKa = 11.84YY84 pKa = 8.07AQNVAQYY91 pKa = 9.57CGTEE95 pKa = 4.27VYY97 pKa = 10.26VVGMSRR103 pKa = 11.84SDD105 pKa = 3.13QRR107 pKa = 11.84KK108 pKa = 9.02DD109 pKa = 3.36MKK111 pKa = 10.79GSRR114 pKa = 11.84QWYY117 pKa = 4.31WTKK120 pKa = 11.1DD121 pKa = 3.23VNAANRR127 pKa = 11.84NDD129 pKa = 3.96RR130 pKa = 11.84PGHH133 pKa = 5.58RR134 pKa = 11.84DD135 pKa = 2.71IRR137 pKa = 11.84FLCDD141 pKa = 2.45VDD143 pKa = 5.16YY144 pKa = 11.08YY145 pKa = 11.83VDD147 pKa = 3.79MPAMLVAEE155 pKa = 5.23AKK157 pKa = 10.01PVLLYY162 pKa = 10.21TVVPEE167 pKa = 4.32EE168 pKa = 4.34ATSSANDD175 pKa = 3.02EE176 pKa = 4.23SAFYY180 pKa = 10.78FEE182 pKa = 5.82ADD184 pKa = 3.08GSLRR188 pKa = 11.84TMVAGGGGYY197 pKa = 7.35SHH199 pKa = 7.39CIWDD203 pKa = 3.7YY204 pKa = 11.32AADD207 pKa = 3.63SFLVVKK213 pKa = 9.17KK214 pKa = 10.28ICGVPIRR221 pKa = 11.84AVAYY225 pKa = 9.43AVEE228 pKa = 4.39RR229 pKa = 11.84KK230 pKa = 8.59QVGKK234 pKa = 9.65HH235 pKa = 4.19RR236 pKa = 11.84QMVLLAPVRR245 pKa = 11.84AFSGLAAILAYY256 pKa = 10.69LLLEE260 pKa = 4.32TKK262 pKa = 8.77EE263 pKa = 3.97LRR265 pKa = 11.84RR266 pKa = 11.84FNPIVEE272 pKa = 4.3AGGEE276 pKa = 4.03KK277 pKa = 9.88FVRR280 pKa = 11.84FNVMSSDD287 pKa = 3.32GALMVTTARR296 pKa = 11.84PGSSLCATVSCADD309 pKa = 4.13DD310 pKa = 4.23DD311 pKa = 5.66AIATVARR318 pKa = 11.84LGSTNLMLPTTASWVKK334 pKa = 10.33DD335 pKa = 3.94RR336 pKa = 11.84PAAAVLTDD344 pKa = 3.36YY345 pKa = 10.84HH346 pKa = 6.78RR347 pKa = 11.84KK348 pKa = 9.29CGKK351 pKa = 8.57KK352 pKa = 10.21AKK354 pKa = 9.67FVVYY358 pKa = 9.47PVEE361 pKa = 4.05QGVRR365 pKa = 11.84AYY367 pKa = 10.17QYY369 pKa = 11.21KK370 pKa = 10.21PDD372 pKa = 4.42EE373 pKa = 4.71FDD375 pKa = 2.84QAARR379 pKa = 11.84PKK381 pKa = 9.8LQAFMSPIVHH391 pKa = 6.5GAFAPVLNRR400 pKa = 11.84AGEE403 pKa = 4.13EE404 pKa = 3.93RR405 pKa = 11.84CVEE408 pKa = 3.86GRR410 pKa = 11.84INSLRR415 pKa = 11.84KK416 pKa = 9.65PEE418 pKa = 3.88PHH420 pKa = 6.95PSNFRR425 pKa = 11.84DD426 pKa = 3.28RR427 pKa = 11.84CIDD430 pKa = 3.4EE431 pKa = 4.59FADD434 pKa = 3.92LVVSGMLLEE443 pKa = 4.38PVCFEE448 pKa = 4.32VVNSKK453 pKa = 7.44QTRR456 pKa = 11.84STQKK460 pKa = 10.64LSIAKK465 pKa = 9.8AVLTGKK471 pKa = 9.85FRR473 pKa = 11.84QAVLKK478 pKa = 10.86CFGKK482 pKa = 10.66AEE484 pKa = 4.58AYY486 pKa = 9.68PDD488 pKa = 3.53VKK490 pKa = 10.81DD491 pKa = 3.85PRR493 pKa = 11.84NISQYY498 pKa = 11.4NDD500 pKa = 3.26ADD502 pKa = 4.14KK503 pKa = 11.63LDD505 pKa = 3.67MATFALALSEE515 pKa = 4.67HH516 pKa = 6.08MKK518 pKa = 9.62QFKK521 pKa = 9.69WYY523 pKa = 10.11GPGKK527 pKa = 8.81TPLEE531 pKa = 3.85IANRR535 pKa = 11.84VVEE538 pKa = 4.43ICQAADD544 pKa = 3.41FVNISDD550 pKa = 3.79YY551 pKa = 11.35HH552 pKa = 8.03RR553 pKa = 11.84MDD555 pKa = 3.0GTITYY560 pKa = 7.32TLRR563 pKa = 11.84RR564 pKa = 11.84VEE566 pKa = 4.33RR567 pKa = 11.84AVCMKK572 pKa = 10.8AFANHH577 pKa = 5.83SACLNEE583 pKa = 3.97LLKK586 pKa = 11.17RR587 pKa = 11.84NVDD590 pKa = 3.34NVGYY594 pKa = 9.55LPHH597 pKa = 6.74GTTFNQGSSHH607 pKa = 6.79GSGCSATSLFQTLRR621 pKa = 11.84AAFNAYY627 pKa = 10.22LGFRR631 pKa = 11.84HH632 pKa = 5.86TRR634 pKa = 11.84NQAGRR639 pKa = 11.84MFSPKK644 pKa = 9.86EE645 pKa = 3.72AFASLGIHH653 pKa = 6.81LGDD656 pKa = 5.87DD657 pKa = 3.98GLDD660 pKa = 3.49GNLPIGSHH668 pKa = 4.91QWASKK673 pKa = 7.08QTGLILEE680 pKa = 4.38AALVYY685 pKa = 10.21RR686 pKa = 11.84GYY688 pKa = 10.82RR689 pKa = 11.84GVNFLARR696 pKa = 11.84YY697 pKa = 7.47YY698 pKa = 10.9SPEE701 pKa = 3.69VWQGCPDD708 pKa = 3.63SMCDD712 pKa = 4.07FKK714 pKa = 11.42RR715 pKa = 11.84QISKK719 pKa = 9.63FHH721 pKa = 5.25TTVRR725 pKa = 11.84LPSNVTPEE733 pKa = 3.87QKK735 pKa = 10.43LVEE738 pKa = 4.08KK739 pKa = 10.74SMSYY743 pKa = 10.74VATDD747 pKa = 3.02GNTPVIGQLCKK758 pKa = 10.16RR759 pKa = 11.84VLLLSSLRR767 pKa = 11.84PRR769 pKa = 11.84TLFGLGNWWSQFDD782 pKa = 5.25DD783 pKa = 4.0SVQFPNNNVGGWMDD797 pKa = 3.73VEE799 pKa = 4.73IDD801 pKa = 3.55HH802 pKa = 6.44QFPEE806 pKa = 4.27FDD808 pKa = 4.17RR809 pKa = 11.84NLFDD813 pKa = 3.38KK814 pKa = 10.63WLDD817 pKa = 3.71SVQKK821 pKa = 11.19AEE823 pKa = 4.9DD824 pKa = 3.93LLSAPLCAEE833 pKa = 4.92PKK835 pKa = 10.07PPTPGCVAVVVDD847 pKa = 4.85EE848 pKa = 4.94EE849 pKa = 4.71IIPPRR854 pKa = 11.84QDD856 pKa = 2.98VEE858 pKa = 4.28PARR861 pKa = 11.84SSSPPRR867 pKa = 11.84RR868 pKa = 11.84TPLRR872 pKa = 11.84PTRR875 pKa = 11.84YY876 pKa = 8.22KK877 pKa = 9.89VTVARR882 pKa = 11.84EE883 pKa = 3.81KK884 pKa = 10.51ATLLNPPKK892 pKa = 9.87PRR894 pKa = 11.84RR895 pKa = 11.84ATAA898 pKa = 3.35
Molecular weight: 99.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8BET0|Q8BET0_9VIRU RNA replicase OS=Sclerophthora macrospora virus A OX=191289 PE=3 SV=1
MM1 pKa = 7.52NYY3 pKa = 10.16SSGMSTTSGTSPTGRR18 pKa = 11.84HH19 pKa = 6.01LIKK22 pKa = 10.33EE23 pKa = 4.24VRR25 pKa = 11.84TDD27 pKa = 3.41QAVQQPASSKK37 pKa = 9.96PSEE40 pKa = 4.1LRR42 pKa = 11.84LTHH45 pKa = 6.74ILDD48 pKa = 3.62SGTRR52 pKa = 11.84GTKK55 pKa = 9.93RR56 pKa = 11.84AEE58 pKa = 4.04CSPLRR63 pKa = 11.84RR64 pKa = 11.84PLQVLASISVTTVSMVTYY82 pKa = 9.99PSDD85 pKa = 3.56LTSGRR90 pKa = 11.84QNRR93 pKa = 11.84QGSYY97 pKa = 7.2WKK99 pKa = 10.6RR100 pKa = 11.84PWCTEE105 pKa = 4.02AIEE108 pKa = 5.14EE109 pKa = 4.52SISWHH114 pKa = 4.63ATIRR118 pKa = 11.84PKK120 pKa = 10.52SGKK123 pKa = 9.88GVLIVCVTSRR133 pKa = 11.84DD134 pKa = 3.46RR135 pKa = 11.84SRR137 pKa = 11.84SSIQRR142 pKa = 11.84FAYY145 pKa = 10.72LLTSRR150 pKa = 11.84LNKK153 pKa = 9.26SWSRR157 pKa = 11.84NPCPMWQPMEE167 pKa = 3.98IHH169 pKa = 6.64PLSVSFARR177 pKa = 11.84EE178 pKa = 4.24CFCSHH183 pKa = 6.99PCAPEE188 pKa = 4.14LFLDD192 pKa = 4.08SVIGGLSSTIPYY204 pKa = 9.96SSPTIMLEE212 pKa = 4.58DD213 pKa = 2.93GWMWKK218 pKa = 10.41LIISSQNSTEE228 pKa = 3.91ISSTSGWIQFKK239 pKa = 10.46KK240 pKa = 10.48RR241 pKa = 11.84RR242 pKa = 11.84TCFQLHH248 pKa = 5.34YY249 pKa = 10.2VLNPSHH255 pKa = 5.94QHH257 pKa = 6.68LDD259 pKa = 3.56ALQSWLTRR267 pKa = 11.84RR268 pKa = 11.84LSLHH272 pKa = 6.87DD273 pKa = 3.89RR274 pKa = 11.84MLSQHH279 pKa = 5.5EE280 pKa = 3.95ALRR283 pKa = 11.84RR284 pKa = 11.84LGEE287 pKa = 3.91HH288 pKa = 6.64RR289 pKa = 4.27
MM1 pKa = 7.52NYY3 pKa = 10.16SSGMSTTSGTSPTGRR18 pKa = 11.84HH19 pKa = 6.01LIKK22 pKa = 10.33EE23 pKa = 4.24VRR25 pKa = 11.84TDD27 pKa = 3.41QAVQQPASSKK37 pKa = 9.96PSEE40 pKa = 4.1LRR42 pKa = 11.84LTHH45 pKa = 6.74ILDD48 pKa = 3.62SGTRR52 pKa = 11.84GTKK55 pKa = 9.93RR56 pKa = 11.84AEE58 pKa = 4.04CSPLRR63 pKa = 11.84RR64 pKa = 11.84PLQVLASISVTTVSMVTYY82 pKa = 9.99PSDD85 pKa = 3.56LTSGRR90 pKa = 11.84QNRR93 pKa = 11.84QGSYY97 pKa = 7.2WKK99 pKa = 10.6RR100 pKa = 11.84PWCTEE105 pKa = 4.02AIEE108 pKa = 5.14EE109 pKa = 4.52SISWHH114 pKa = 4.63ATIRR118 pKa = 11.84PKK120 pKa = 10.52SGKK123 pKa = 9.88GVLIVCVTSRR133 pKa = 11.84DD134 pKa = 3.46RR135 pKa = 11.84SRR137 pKa = 11.84SSIQRR142 pKa = 11.84FAYY145 pKa = 10.72LLTSRR150 pKa = 11.84LNKK153 pKa = 9.26SWSRR157 pKa = 11.84NPCPMWQPMEE167 pKa = 3.98IHH169 pKa = 6.64PLSVSFARR177 pKa = 11.84EE178 pKa = 4.24CFCSHH183 pKa = 6.99PCAPEE188 pKa = 4.14LFLDD192 pKa = 4.08SVIGGLSSTIPYY204 pKa = 9.96SSPTIMLEE212 pKa = 4.58DD213 pKa = 2.93GWMWKK218 pKa = 10.41LIISSQNSTEE228 pKa = 3.91ISSTSGWIQFKK239 pKa = 10.46KK240 pKa = 10.48RR241 pKa = 11.84RR242 pKa = 11.84TCFQLHH248 pKa = 5.34YY249 pKa = 10.2VLNPSHH255 pKa = 5.94QHH257 pKa = 6.68LDD259 pKa = 3.56ALQSWLTRR267 pKa = 11.84RR268 pKa = 11.84LSLHH272 pKa = 6.87DD273 pKa = 3.89RR274 pKa = 11.84MLSQHH279 pKa = 5.5EE280 pKa = 3.95ALRR283 pKa = 11.84RR284 pKa = 11.84LGEE287 pKa = 3.91HH288 pKa = 6.64RR289 pKa = 4.27
Molecular weight: 32.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1609 |
289 |
898 |
536.3 |
58.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.136 ± 1.968 | 2.362 ± 0.488 |
4.475 ± 1.081 | 4.102 ± 0.52 |
3.543 ± 0.569 | 6.712 ± 1.085 |
2.113 ± 0.73 | 3.853 ± 0.722 |
4.537 ± 0.545 | 8.515 ± 0.432 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.362 ± 0.152 | 3.915 ± 0.921 |
5.718 ± 0.209 | 3.729 ± 0.421 |
6.464 ± 1.408 | 9.012 ± 2.41 |
6.401 ± 0.882 | 8.142 ± 1.629 |
1.678 ± 0.541 | 3.232 ± 0.455 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
