
Lucheng Rn rat coronavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Luchacovirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
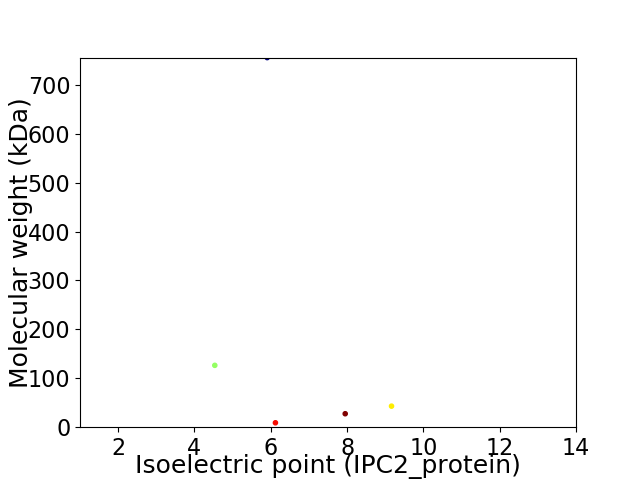
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L7HJ82|A0A1L7HJ82_9ALPC Spike glycoprotein OS=Lucheng Rn rat coronavirus OX=1508224 PE=4 SV=1
MM1 pKa = 7.39YY2 pKa = 10.37FFLLLLFVSADD13 pKa = 3.48AAIQTCPAPGNVNLDD28 pKa = 3.08ISKK31 pKa = 10.61LYY33 pKa = 10.88YY34 pKa = 9.2GTQASINATFQVVQVLPQVPWKK56 pKa = 10.45CNSYY60 pKa = 11.66SNGPSNKK67 pKa = 9.24FNGIGVFVDD76 pKa = 4.33LASAQHH82 pKa = 5.57SWHH85 pKa = 6.3LFVYY89 pKa = 10.33PSMPTNKK96 pKa = 8.32TWILSWADD104 pKa = 3.06THH106 pKa = 6.73TSFEE110 pKa = 4.36HH111 pKa = 6.53GSVISYY117 pKa = 8.02VQICKK122 pKa = 10.34YY123 pKa = 9.0PSNVVTDD130 pKa = 3.52ITNGNGCHH138 pKa = 6.14TNMAGPGATTCDD150 pKa = 3.88VILSSPLEE158 pKa = 4.14CVLNRR163 pKa = 11.84TYY165 pKa = 10.65SQQYY169 pKa = 10.18AGVSYY174 pKa = 8.6ITWYY178 pKa = 9.76NDD180 pKa = 3.4HH181 pKa = 7.25IIASIQGEE189 pKa = 4.73VFTFDD194 pKa = 3.48IGEE197 pKa = 4.11VLQWSNFSAFCGTGNKK213 pKa = 10.04CGFSYY218 pKa = 9.67ATTLSEE224 pKa = 3.78WLVRR228 pKa = 11.84TDD230 pKa = 3.58NDD232 pKa = 3.71GTVIDD237 pKa = 4.82YY238 pKa = 7.72VICDD242 pKa = 3.76TDD244 pKa = 4.17FEE246 pKa = 4.75SQLKK250 pKa = 8.56CKK252 pKa = 10.72NMVFEE257 pKa = 4.74LTPAVYY263 pKa = 10.25SGSAVEE269 pKa = 4.29LQSAIYY275 pKa = 9.56YY276 pKa = 9.33VSNEE280 pKa = 4.29LPDD283 pKa = 4.28CDD285 pKa = 5.55FSFADD290 pKa = 3.78MFMDD294 pKa = 4.14GTGNFEE300 pKa = 3.64GLRR303 pKa = 11.84RR304 pKa = 11.84HH305 pKa = 5.69VFSNCWVNYY314 pKa = 6.46TAWFACADD322 pKa = 4.31DD323 pKa = 4.03YY324 pKa = 12.04SCIIFNAIFAEE335 pKa = 4.2VRR337 pKa = 11.84YY338 pKa = 10.12KK339 pKa = 10.88LSQPDD344 pKa = 3.63GLVNPFIKK352 pKa = 10.71CNGLDD357 pKa = 3.78LYY359 pKa = 11.0SITKK363 pKa = 8.93GCSSGFVLRR372 pKa = 11.84YY373 pKa = 9.33QLYY376 pKa = 9.87ANGSFDD382 pKa = 4.02VNSYY386 pKa = 9.25TPDD389 pKa = 3.21YY390 pKa = 9.16MEE392 pKa = 4.66CFGYY396 pKa = 9.87FALYY400 pKa = 9.58NGYY403 pKa = 10.4VIYY406 pKa = 10.4NAKK409 pKa = 9.93FVSKK413 pKa = 10.42GLTVCVVQPVEE424 pKa = 4.53PEE426 pKa = 3.77LDD428 pKa = 3.41VCKK431 pKa = 10.46SYY433 pKa = 10.68TIDD436 pKa = 3.13GVTFQGILRR445 pKa = 11.84NTTHH449 pKa = 6.99QIDD452 pKa = 3.85SFHH455 pKa = 6.93NILYY459 pKa = 9.35YY460 pKa = 10.98GDD462 pKa = 3.43MVSYY466 pKa = 10.32VRR468 pKa = 11.84IRR470 pKa = 11.84GVVYY474 pKa = 10.34AVEE477 pKa = 4.24SCNRR481 pKa = 11.84FYY483 pKa = 11.67YY484 pKa = 10.69SVFKK488 pKa = 9.96TLSAIGYY495 pKa = 8.24LYY497 pKa = 10.93SGATCDD503 pKa = 3.72STDD506 pKa = 2.76VTTFMTKK513 pKa = 9.79ARR515 pKa = 11.84ATTFVDD521 pKa = 3.31SSLGCFIDD529 pKa = 4.58VSVTDD534 pKa = 4.99GNYY537 pKa = 9.43TDD539 pKa = 4.64CLNPIGNGFCVDD551 pKa = 3.72VNVTGQPVVGNIFIQTHH568 pKa = 5.14DD569 pKa = 3.41TDD571 pKa = 3.78YY572 pKa = 11.58ARR574 pKa = 11.84PILTAQQIEE583 pKa = 4.61LPIDD587 pKa = 3.45HH588 pKa = 6.39YY589 pKa = 11.93VSVKK593 pKa = 9.75EE594 pKa = 4.06QFIQTSTPKK603 pKa = 10.22FDD605 pKa = 3.87VDD607 pKa = 3.72CEE609 pKa = 4.22RR610 pKa = 11.84YY611 pKa = 9.46ICDD614 pKa = 3.08VSSDD618 pKa = 3.63CRR620 pKa = 11.84EE621 pKa = 4.04LLVKK625 pKa = 10.85YY626 pKa = 10.33GGYY629 pKa = 9.55CSKK632 pKa = 10.48ILADD636 pKa = 3.97IKK638 pKa = 10.99SSSIQLDD645 pKa = 3.72YY646 pKa = 11.29QILGLYY652 pKa = 7.6KK653 pKa = 10.11TLAVDD658 pKa = 4.82FKK660 pKa = 11.66VPDD663 pKa = 3.36IDD665 pKa = 5.12FGDD668 pKa = 4.56FNFSMYY674 pKa = 9.85MSEE677 pKa = 4.07ANGRR681 pKa = 11.84SFIEE685 pKa = 4.8DD686 pKa = 3.74LLFDD690 pKa = 5.23KK691 pKa = 10.72IVTTGPGFYY700 pKa = 9.77QDD702 pKa = 5.25YY703 pKa = 10.36YY704 pKa = 11.41DD705 pKa = 4.07CKK707 pKa = 11.08KK708 pKa = 10.59MNLQDD713 pKa = 4.03LTCKK717 pKa = 10.15QYY719 pKa = 11.27YY720 pKa = 8.62NGIMVIPPVMDD731 pKa = 3.47DD732 pKa = 3.41TLITFWSSAVAGSMTAGLFGGQAGMVSWTVALAGRR767 pKa = 11.84LNALGVMQDD776 pKa = 3.55ALVEE780 pKa = 4.24DD781 pKa = 4.41VNKK784 pKa = 10.3LANGFNNLTQYY795 pKa = 10.95VSDD798 pKa = 3.99GFKK801 pKa = 8.42TTSQALSTIQAVVNNNAQQVSQLVQGLSEE830 pKa = 4.2NFGAISNNFALIAEE844 pKa = 4.1RR845 pKa = 11.84LEE847 pKa = 4.47RR848 pKa = 11.84IEE850 pKa = 5.82AAMQMDD856 pKa = 4.41RR857 pKa = 11.84LINGRR862 pKa = 11.84MNILQNFVTNYY873 pKa = 9.51KK874 pKa = 10.59LSISEE879 pKa = 4.69LKK881 pKa = 10.33SQQALAQSLINEE893 pKa = 4.31CVYY896 pKa = 10.93AQSSRR901 pKa = 11.84NGFCGDD907 pKa = 3.51GLHH910 pKa = 6.68LFSLMQRR917 pKa = 11.84APDD920 pKa = 4.2GIMFFHH926 pKa = 6.52YY927 pKa = 9.1TLKK930 pKa = 10.61PNNTIIVEE938 pKa = 4.32TTPGLCLSNDD948 pKa = 3.04VCIAPKK954 pKa = 10.5DD955 pKa = 3.73GLFVRR960 pKa = 11.84LSTARR965 pKa = 11.84DD966 pKa = 3.78TDD968 pKa = 3.13WHH970 pKa = 5.32FTTRR974 pKa = 11.84NRR976 pKa = 11.84YY977 pKa = 8.21SPEE980 pKa = 4.89PITVNNTLTISGGVNFTVVNSTIDD1004 pKa = 4.17GIEE1007 pKa = 4.13PPANPSFDD1015 pKa = 3.75EE1016 pKa = 4.55EE1017 pKa = 4.15FAEE1020 pKa = 4.83LYY1022 pKa = 11.03KK1023 pKa = 11.06NVTLEE1028 pKa = 4.5LEE1030 pKa = 4.02QLKK1033 pKa = 10.79NISFDD1038 pKa = 3.78PEE1040 pKa = 4.25MLNLTYY1046 pKa = 10.84YY1047 pKa = 10.11IDD1049 pKa = 4.39RR1050 pKa = 11.84LDD1052 pKa = 3.88EE1053 pKa = 4.2LATNVSQLHH1062 pKa = 5.6VDD1064 pKa = 3.09VSEE1067 pKa = 4.22FNKK1070 pKa = 9.68FVQYY1074 pKa = 10.55IKK1076 pKa = 10.02WPWYY1080 pKa = 7.76VWLAIFLVLVLFSFLMLWCCCATGCCGCCGLCGAACNGCCTKK1122 pKa = 10.18PQPIEE1127 pKa = 3.79FEE1129 pKa = 4.31KK1130 pKa = 11.18VHH1132 pKa = 5.12VQQ1134 pKa = 3.25
MM1 pKa = 7.39YY2 pKa = 10.37FFLLLLFVSADD13 pKa = 3.48AAIQTCPAPGNVNLDD28 pKa = 3.08ISKK31 pKa = 10.61LYY33 pKa = 10.88YY34 pKa = 9.2GTQASINATFQVVQVLPQVPWKK56 pKa = 10.45CNSYY60 pKa = 11.66SNGPSNKK67 pKa = 9.24FNGIGVFVDD76 pKa = 4.33LASAQHH82 pKa = 5.57SWHH85 pKa = 6.3LFVYY89 pKa = 10.33PSMPTNKK96 pKa = 8.32TWILSWADD104 pKa = 3.06THH106 pKa = 6.73TSFEE110 pKa = 4.36HH111 pKa = 6.53GSVISYY117 pKa = 8.02VQICKK122 pKa = 10.34YY123 pKa = 9.0PSNVVTDD130 pKa = 3.52ITNGNGCHH138 pKa = 6.14TNMAGPGATTCDD150 pKa = 3.88VILSSPLEE158 pKa = 4.14CVLNRR163 pKa = 11.84TYY165 pKa = 10.65SQQYY169 pKa = 10.18AGVSYY174 pKa = 8.6ITWYY178 pKa = 9.76NDD180 pKa = 3.4HH181 pKa = 7.25IIASIQGEE189 pKa = 4.73VFTFDD194 pKa = 3.48IGEE197 pKa = 4.11VLQWSNFSAFCGTGNKK213 pKa = 10.04CGFSYY218 pKa = 9.67ATTLSEE224 pKa = 3.78WLVRR228 pKa = 11.84TDD230 pKa = 3.58NDD232 pKa = 3.71GTVIDD237 pKa = 4.82YY238 pKa = 7.72VICDD242 pKa = 3.76TDD244 pKa = 4.17FEE246 pKa = 4.75SQLKK250 pKa = 8.56CKK252 pKa = 10.72NMVFEE257 pKa = 4.74LTPAVYY263 pKa = 10.25SGSAVEE269 pKa = 4.29LQSAIYY275 pKa = 9.56YY276 pKa = 9.33VSNEE280 pKa = 4.29LPDD283 pKa = 4.28CDD285 pKa = 5.55FSFADD290 pKa = 3.78MFMDD294 pKa = 4.14GTGNFEE300 pKa = 3.64GLRR303 pKa = 11.84RR304 pKa = 11.84HH305 pKa = 5.69VFSNCWVNYY314 pKa = 6.46TAWFACADD322 pKa = 4.31DD323 pKa = 4.03YY324 pKa = 12.04SCIIFNAIFAEE335 pKa = 4.2VRR337 pKa = 11.84YY338 pKa = 10.12KK339 pKa = 10.88LSQPDD344 pKa = 3.63GLVNPFIKK352 pKa = 10.71CNGLDD357 pKa = 3.78LYY359 pKa = 11.0SITKK363 pKa = 8.93GCSSGFVLRR372 pKa = 11.84YY373 pKa = 9.33QLYY376 pKa = 9.87ANGSFDD382 pKa = 4.02VNSYY386 pKa = 9.25TPDD389 pKa = 3.21YY390 pKa = 9.16MEE392 pKa = 4.66CFGYY396 pKa = 9.87FALYY400 pKa = 9.58NGYY403 pKa = 10.4VIYY406 pKa = 10.4NAKK409 pKa = 9.93FVSKK413 pKa = 10.42GLTVCVVQPVEE424 pKa = 4.53PEE426 pKa = 3.77LDD428 pKa = 3.41VCKK431 pKa = 10.46SYY433 pKa = 10.68TIDD436 pKa = 3.13GVTFQGILRR445 pKa = 11.84NTTHH449 pKa = 6.99QIDD452 pKa = 3.85SFHH455 pKa = 6.93NILYY459 pKa = 9.35YY460 pKa = 10.98GDD462 pKa = 3.43MVSYY466 pKa = 10.32VRR468 pKa = 11.84IRR470 pKa = 11.84GVVYY474 pKa = 10.34AVEE477 pKa = 4.24SCNRR481 pKa = 11.84FYY483 pKa = 11.67YY484 pKa = 10.69SVFKK488 pKa = 9.96TLSAIGYY495 pKa = 8.24LYY497 pKa = 10.93SGATCDD503 pKa = 3.72STDD506 pKa = 2.76VTTFMTKK513 pKa = 9.79ARR515 pKa = 11.84ATTFVDD521 pKa = 3.31SSLGCFIDD529 pKa = 4.58VSVTDD534 pKa = 4.99GNYY537 pKa = 9.43TDD539 pKa = 4.64CLNPIGNGFCVDD551 pKa = 3.72VNVTGQPVVGNIFIQTHH568 pKa = 5.14DD569 pKa = 3.41TDD571 pKa = 3.78YY572 pKa = 11.58ARR574 pKa = 11.84PILTAQQIEE583 pKa = 4.61LPIDD587 pKa = 3.45HH588 pKa = 6.39YY589 pKa = 11.93VSVKK593 pKa = 9.75EE594 pKa = 4.06QFIQTSTPKK603 pKa = 10.22FDD605 pKa = 3.87VDD607 pKa = 3.72CEE609 pKa = 4.22RR610 pKa = 11.84YY611 pKa = 9.46ICDD614 pKa = 3.08VSSDD618 pKa = 3.63CRR620 pKa = 11.84EE621 pKa = 4.04LLVKK625 pKa = 10.85YY626 pKa = 10.33GGYY629 pKa = 9.55CSKK632 pKa = 10.48ILADD636 pKa = 3.97IKK638 pKa = 10.99SSSIQLDD645 pKa = 3.72YY646 pKa = 11.29QILGLYY652 pKa = 7.6KK653 pKa = 10.11TLAVDD658 pKa = 4.82FKK660 pKa = 11.66VPDD663 pKa = 3.36IDD665 pKa = 5.12FGDD668 pKa = 4.56FNFSMYY674 pKa = 9.85MSEE677 pKa = 4.07ANGRR681 pKa = 11.84SFIEE685 pKa = 4.8DD686 pKa = 3.74LLFDD690 pKa = 5.23KK691 pKa = 10.72IVTTGPGFYY700 pKa = 9.77QDD702 pKa = 5.25YY703 pKa = 10.36YY704 pKa = 11.41DD705 pKa = 4.07CKK707 pKa = 11.08KK708 pKa = 10.59MNLQDD713 pKa = 4.03LTCKK717 pKa = 10.15QYY719 pKa = 11.27YY720 pKa = 8.62NGIMVIPPVMDD731 pKa = 3.47DD732 pKa = 3.41TLITFWSSAVAGSMTAGLFGGQAGMVSWTVALAGRR767 pKa = 11.84LNALGVMQDD776 pKa = 3.55ALVEE780 pKa = 4.24DD781 pKa = 4.41VNKK784 pKa = 10.3LANGFNNLTQYY795 pKa = 10.95VSDD798 pKa = 3.99GFKK801 pKa = 8.42TTSQALSTIQAVVNNNAQQVSQLVQGLSEE830 pKa = 4.2NFGAISNNFALIAEE844 pKa = 4.1RR845 pKa = 11.84LEE847 pKa = 4.47RR848 pKa = 11.84IEE850 pKa = 5.82AAMQMDD856 pKa = 4.41RR857 pKa = 11.84LINGRR862 pKa = 11.84MNILQNFVTNYY873 pKa = 9.51KK874 pKa = 10.59LSISEE879 pKa = 4.69LKK881 pKa = 10.33SQQALAQSLINEE893 pKa = 4.31CVYY896 pKa = 10.93AQSSRR901 pKa = 11.84NGFCGDD907 pKa = 3.51GLHH910 pKa = 6.68LFSLMQRR917 pKa = 11.84APDD920 pKa = 4.2GIMFFHH926 pKa = 6.52YY927 pKa = 9.1TLKK930 pKa = 10.61PNNTIIVEE938 pKa = 4.32TTPGLCLSNDD948 pKa = 3.04VCIAPKK954 pKa = 10.5DD955 pKa = 3.73GLFVRR960 pKa = 11.84LSTARR965 pKa = 11.84DD966 pKa = 3.78TDD968 pKa = 3.13WHH970 pKa = 5.32FTTRR974 pKa = 11.84NRR976 pKa = 11.84YY977 pKa = 8.21SPEE980 pKa = 4.89PITVNNTLTISGGVNFTVVNSTIDD1004 pKa = 4.17GIEE1007 pKa = 4.13PPANPSFDD1015 pKa = 3.75EE1016 pKa = 4.55EE1017 pKa = 4.15FAEE1020 pKa = 4.83LYY1022 pKa = 11.03KK1023 pKa = 11.06NVTLEE1028 pKa = 4.5LEE1030 pKa = 4.02QLKK1033 pKa = 10.79NISFDD1038 pKa = 3.78PEE1040 pKa = 4.25MLNLTYY1046 pKa = 10.84YY1047 pKa = 10.11IDD1049 pKa = 4.39RR1050 pKa = 11.84LDD1052 pKa = 3.88EE1053 pKa = 4.2LATNVSQLHH1062 pKa = 5.6VDD1064 pKa = 3.09VSEE1067 pKa = 4.22FNKK1070 pKa = 9.68FVQYY1074 pKa = 10.55IKK1076 pKa = 10.02WPWYY1080 pKa = 7.76VWLAIFLVLVLFSFLMLWCCCATGCCGCCGLCGAACNGCCTKK1122 pKa = 10.18PQPIEE1127 pKa = 3.79FEE1129 pKa = 4.31KK1130 pKa = 11.18VHH1132 pKa = 5.12VQQ1134 pKa = 3.25
Molecular weight: 126.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L7HJ65|A0A1L7HJ65_9ALPC Membrane protein OS=Lucheng Rn rat coronavirus OX=1508224 GN=M PE=3 SV=1
MM1 pKa = 7.45SSNVSWADD9 pKa = 3.15QVEE12 pKa = 4.7TVNRR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84SRR21 pKa = 11.84SRR23 pKa = 11.84GRR25 pKa = 11.84SQNRR29 pKa = 11.84TNASNPLSWFTSIIDD44 pKa = 3.59EE45 pKa = 4.55SNGNFISLMPHH56 pKa = 5.84SGVPTGMGTAAQQCGYY72 pKa = 8.99WYY74 pKa = 9.86RR75 pKa = 11.84APTVYY80 pKa = 9.83QVRR83 pKa = 11.84RR84 pKa = 11.84GKK86 pKa = 10.02RR87 pKa = 11.84VPLPPVWYY95 pKa = 8.96FYY97 pKa = 11.33FLGTGPHH104 pKa = 6.53ANAAYY109 pKa = 7.11GTAMDD114 pKa = 3.85GVFWVKK120 pKa = 9.53TKK122 pKa = 10.78NGQIDD127 pKa = 3.96PKK129 pKa = 9.07STKK132 pKa = 10.51ALGVRR137 pKa = 11.84DD138 pKa = 3.77SGTDD142 pKa = 3.25PRR144 pKa = 11.84RR145 pKa = 11.84ANIPNLPEE153 pKa = 3.99GLRR156 pKa = 11.84VNVPNASRR164 pKa = 11.84PQSRR168 pKa = 11.84AQSQTRR174 pKa = 11.84SQNTSRR180 pKa = 11.84ASSVSRR186 pKa = 11.84NGSRR190 pKa = 11.84APSVDD195 pKa = 3.03RR196 pKa = 11.84TKK198 pKa = 11.28EE199 pKa = 3.74DD200 pKa = 3.04LKK202 pKa = 11.41AVVSQLLAEE211 pKa = 4.39MGISKK216 pKa = 10.34NSKK219 pKa = 7.23QTQNQKK225 pKa = 9.11KK226 pKa = 9.71QKK228 pKa = 9.96GSTPAATPHH237 pKa = 6.75PNQDD241 pKa = 3.55GKK243 pKa = 9.81PVWKK247 pKa = 10.25KK248 pKa = 10.68KK249 pKa = 9.19PNKK252 pKa = 9.71EE253 pKa = 3.8EE254 pKa = 3.86TVAQCFGPRR263 pKa = 11.84SDD265 pKa = 3.88NKK267 pKa = 10.55NFGDD271 pKa = 4.76AEE273 pKa = 4.06FLRR276 pKa = 11.84LGVDD280 pKa = 3.38DD281 pKa = 5.19PRR283 pKa = 11.84FKK285 pKa = 10.14TASYY289 pKa = 8.67YY290 pKa = 11.24APGAAASLFDD300 pKa = 4.33SMVTVSDD307 pKa = 4.08GPDD310 pKa = 2.93GKK312 pKa = 10.96KK313 pKa = 9.72RR314 pKa = 11.84VTFHH318 pKa = 5.24TTIEE322 pKa = 4.02VDD324 pKa = 3.14PTKK327 pKa = 10.65PGFEE331 pKa = 4.09VFLAQIDD338 pKa = 4.16AFKK341 pKa = 11.0KK342 pKa = 9.97PATFQQTQNFWEE354 pKa = 4.47NQASTQNNISEE365 pKa = 4.68YY366 pKa = 10.63FRR368 pKa = 11.84GTTPGAGGSAVEE380 pKa = 4.19IEE382 pKa = 4.46TFEE385 pKa = 4.26MTDD388 pKa = 3.2EE389 pKa = 4.43TNN391 pKa = 3.0
MM1 pKa = 7.45SSNVSWADD9 pKa = 3.15QVEE12 pKa = 4.7TVNRR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84SRR21 pKa = 11.84SRR23 pKa = 11.84GRR25 pKa = 11.84SQNRR29 pKa = 11.84TNASNPLSWFTSIIDD44 pKa = 3.59EE45 pKa = 4.55SNGNFISLMPHH56 pKa = 5.84SGVPTGMGTAAQQCGYY72 pKa = 8.99WYY74 pKa = 9.86RR75 pKa = 11.84APTVYY80 pKa = 9.83QVRR83 pKa = 11.84RR84 pKa = 11.84GKK86 pKa = 10.02RR87 pKa = 11.84VPLPPVWYY95 pKa = 8.96FYY97 pKa = 11.33FLGTGPHH104 pKa = 6.53ANAAYY109 pKa = 7.11GTAMDD114 pKa = 3.85GVFWVKK120 pKa = 9.53TKK122 pKa = 10.78NGQIDD127 pKa = 3.96PKK129 pKa = 9.07STKK132 pKa = 10.51ALGVRR137 pKa = 11.84DD138 pKa = 3.77SGTDD142 pKa = 3.25PRR144 pKa = 11.84RR145 pKa = 11.84ANIPNLPEE153 pKa = 3.99GLRR156 pKa = 11.84VNVPNASRR164 pKa = 11.84PQSRR168 pKa = 11.84AQSQTRR174 pKa = 11.84SQNTSRR180 pKa = 11.84ASSVSRR186 pKa = 11.84NGSRR190 pKa = 11.84APSVDD195 pKa = 3.03RR196 pKa = 11.84TKK198 pKa = 11.28EE199 pKa = 3.74DD200 pKa = 3.04LKK202 pKa = 11.41AVVSQLLAEE211 pKa = 4.39MGISKK216 pKa = 10.34NSKK219 pKa = 7.23QTQNQKK225 pKa = 9.11KK226 pKa = 9.71QKK228 pKa = 9.96GSTPAATPHH237 pKa = 6.75PNQDD241 pKa = 3.55GKK243 pKa = 9.81PVWKK247 pKa = 10.25KK248 pKa = 10.68KK249 pKa = 9.19PNKK252 pKa = 9.71EE253 pKa = 3.8EE254 pKa = 3.86TVAQCFGPRR263 pKa = 11.84SDD265 pKa = 3.88NKK267 pKa = 10.55NFGDD271 pKa = 4.76AEE273 pKa = 4.06FLRR276 pKa = 11.84LGVDD280 pKa = 3.38DD281 pKa = 5.19PRR283 pKa = 11.84FKK285 pKa = 10.14TASYY289 pKa = 8.67YY290 pKa = 11.24APGAAASLFDD300 pKa = 4.33SMVTVSDD307 pKa = 4.08GPDD310 pKa = 2.93GKK312 pKa = 10.96KK313 pKa = 9.72RR314 pKa = 11.84VTFHH318 pKa = 5.24TTIEE322 pKa = 4.02VDD324 pKa = 3.14PTKK327 pKa = 10.65PGFEE331 pKa = 4.09VFLAQIDD338 pKa = 4.16AFKK341 pKa = 11.0KK342 pKa = 9.97PATFQQTQNFWEE354 pKa = 4.47NQASTQNNISEE365 pKa = 4.68YY366 pKa = 10.63FRR368 pKa = 11.84GTTPGAGGSAVEE380 pKa = 4.19IEE382 pKa = 4.46TFEE385 pKa = 4.26MTDD388 pKa = 3.2EE389 pKa = 4.43TNN391 pKa = 3.0
Molecular weight: 43.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8600 |
78 |
6749 |
1720.0 |
192.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.86 ± 0.499 | 3.674 ± 0.739 |
6.047 ± 0.958 | 3.837 ± 0.266 |
5.895 ± 0.305 | 6.547 ± 0.268 |
1.884 ± 0.332 | 4.86 ± 0.729 |
5.419 ± 0.828 | 8.488 ± 0.977 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.318 | 5.605 ± 0.445 |
3.442 ± 0.694 | 3.058 ± 0.808 |
3.605 ± 0.589 | 7.186 ± 0.662 |
6.035 ± 0.757 | 8.895 ± 0.568 |
1.36 ± 0.443 | 5.128 ± 0.496 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
