
Macaca mulatta feces associated virus 10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus macas6
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
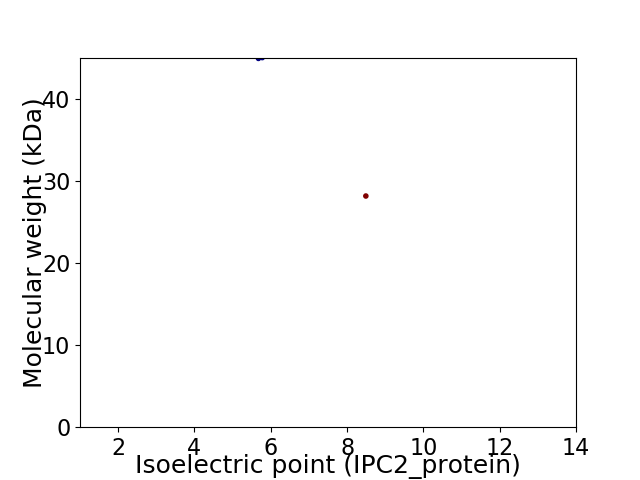
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5PX53|A0A1W5PX53_9VIRU Cap OS=Macaca mulatta feces associated virus 10 OX=2499232 PE=4 SV=1
MM1 pKa = 7.96PILLGMRR8 pKa = 11.84FLCIDD13 pKa = 3.88IGCEE17 pKa = 4.2IMTTTQFASASYY29 pKa = 10.78QEE31 pKa = 5.1VIDD34 pKa = 5.17LSTTGDD40 pKa = 3.45TVSAIGIHH48 pKa = 5.61TPCSATPVDD57 pKa = 4.02MLSGFWRR64 pKa = 11.84QFQKK68 pKa = 10.52VRR70 pKa = 11.84YY71 pKa = 8.55DD72 pKa = 3.44GCSVTLVPAARR83 pKa = 11.84LPVDD87 pKa = 3.59PLGVSYY93 pKa = 10.41EE94 pKa = 4.09AGEE97 pKa = 4.05ATIDD101 pKa = 4.58PRR103 pKa = 11.84DD104 pKa = 3.9LLNPILFHH112 pKa = 6.51GCHH115 pKa = 6.26GNDD118 pKa = 2.85MGYY121 pKa = 9.85ILNQLYY127 pKa = 10.93SGDD130 pKa = 3.77ATISEE135 pKa = 4.09AWMQKK140 pKa = 9.89VGASDD145 pKa = 3.38STEE148 pKa = 4.01QLNMIADD155 pKa = 4.12GAEE158 pKa = 4.38SINTVAEE165 pKa = 3.8ALYY168 pKa = 10.87YY169 pKa = 10.38KK170 pKa = 10.8ALTDD174 pKa = 3.94NTWKK178 pKa = 10.76KK179 pKa = 10.93ADD181 pKa = 3.66PQRR184 pKa = 11.84GFKK187 pKa = 10.51KK188 pKa = 10.36SGLRR192 pKa = 11.84PLVHH196 pKa = 7.23DD197 pKa = 4.74LAVNVPFGVGKK208 pKa = 9.85HH209 pKa = 5.9LGAEE213 pKa = 4.34VTGTSASIQNGRR225 pKa = 11.84FATASPTHH233 pKa = 6.55PAGSGNDD240 pKa = 3.84DD241 pKa = 4.4SIQTSDD247 pKa = 4.72LGFLNTGFNSLGFPDD262 pKa = 4.27TTWTPIIGQNPGKK275 pKa = 10.27LVIGSNPNGIQFMTNRR291 pKa = 11.84LRR293 pKa = 11.84SLGWLDD299 pKa = 3.32TRR301 pKa = 11.84SPLARR306 pKa = 11.84IQTGSSGVTIPSGSRR321 pKa = 11.84AGDD324 pKa = 3.28IGPSTPSGPSRR335 pKa = 11.84SVTDD339 pKa = 3.46VYY341 pKa = 11.64AKK343 pKa = 10.7VIAEE347 pKa = 4.54DD348 pKa = 4.12GNTARR353 pKa = 11.84AVEE356 pKa = 4.03NTIPKK361 pKa = 9.78IFMGMILLPPAYY373 pKa = 8.29KK374 pKa = 9.3TKK376 pKa = 10.4QYY378 pKa = 10.49FRR380 pKa = 11.84MVINHH385 pKa = 6.71RR386 pKa = 11.84FSFAKK391 pKa = 10.18FRR393 pKa = 11.84GISMNNMSTDD403 pKa = 3.77EE404 pKa = 4.24VKK406 pKa = 10.44SAYY409 pKa = 9.98AYY411 pKa = 10.25YY412 pKa = 10.8DD413 pKa = 3.48NYY415 pKa = 11.42SSS417 pKa = 3.69
MM1 pKa = 7.96PILLGMRR8 pKa = 11.84FLCIDD13 pKa = 3.88IGCEE17 pKa = 4.2IMTTTQFASASYY29 pKa = 10.78QEE31 pKa = 5.1VIDD34 pKa = 5.17LSTTGDD40 pKa = 3.45TVSAIGIHH48 pKa = 5.61TPCSATPVDD57 pKa = 4.02MLSGFWRR64 pKa = 11.84QFQKK68 pKa = 10.52VRR70 pKa = 11.84YY71 pKa = 8.55DD72 pKa = 3.44GCSVTLVPAARR83 pKa = 11.84LPVDD87 pKa = 3.59PLGVSYY93 pKa = 10.41EE94 pKa = 4.09AGEE97 pKa = 4.05ATIDD101 pKa = 4.58PRR103 pKa = 11.84DD104 pKa = 3.9LLNPILFHH112 pKa = 6.51GCHH115 pKa = 6.26GNDD118 pKa = 2.85MGYY121 pKa = 9.85ILNQLYY127 pKa = 10.93SGDD130 pKa = 3.77ATISEE135 pKa = 4.09AWMQKK140 pKa = 9.89VGASDD145 pKa = 3.38STEE148 pKa = 4.01QLNMIADD155 pKa = 4.12GAEE158 pKa = 4.38SINTVAEE165 pKa = 3.8ALYY168 pKa = 10.87YY169 pKa = 10.38KK170 pKa = 10.8ALTDD174 pKa = 3.94NTWKK178 pKa = 10.76KK179 pKa = 10.93ADD181 pKa = 3.66PQRR184 pKa = 11.84GFKK187 pKa = 10.51KK188 pKa = 10.36SGLRR192 pKa = 11.84PLVHH196 pKa = 7.23DD197 pKa = 4.74LAVNVPFGVGKK208 pKa = 9.85HH209 pKa = 5.9LGAEE213 pKa = 4.34VTGTSASIQNGRR225 pKa = 11.84FATASPTHH233 pKa = 6.55PAGSGNDD240 pKa = 3.84DD241 pKa = 4.4SIQTSDD247 pKa = 4.72LGFLNTGFNSLGFPDD262 pKa = 4.27TTWTPIIGQNPGKK275 pKa = 10.27LVIGSNPNGIQFMTNRR291 pKa = 11.84LRR293 pKa = 11.84SLGWLDD299 pKa = 3.32TRR301 pKa = 11.84SPLARR306 pKa = 11.84IQTGSSGVTIPSGSRR321 pKa = 11.84AGDD324 pKa = 3.28IGPSTPSGPSRR335 pKa = 11.84SVTDD339 pKa = 3.46VYY341 pKa = 11.64AKK343 pKa = 10.7VIAEE347 pKa = 4.54DD348 pKa = 4.12GNTARR353 pKa = 11.84AVEE356 pKa = 4.03NTIPKK361 pKa = 9.78IFMGMILLPPAYY373 pKa = 8.29KK374 pKa = 9.3TKK376 pKa = 10.4QYY378 pKa = 10.49FRR380 pKa = 11.84MVINHH385 pKa = 6.71RR386 pKa = 11.84FSFAKK391 pKa = 10.18FRR393 pKa = 11.84GISMNNMSTDD403 pKa = 3.77EE404 pKa = 4.24VKK406 pKa = 10.44SAYY409 pKa = 9.98AYY411 pKa = 10.25YY412 pKa = 10.8DD413 pKa = 3.48NYY415 pKa = 11.42SSS417 pKa = 3.69
Molecular weight: 44.91 kDa
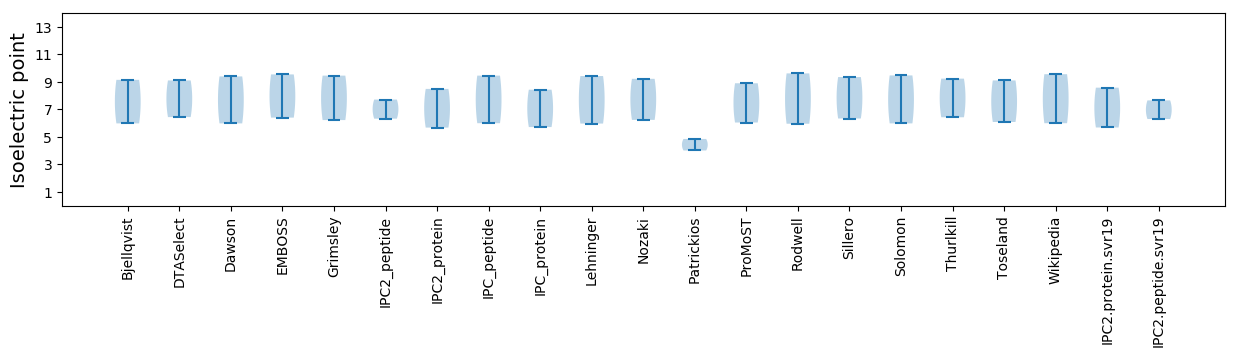
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5PX53|A0A1W5PX53_9VIRU Cap OS=Macaca mulatta feces associated virus 10 OX=2499232 PE=4 SV=1
MM1 pKa = 7.37IMSKK5 pKa = 8.69TFMLTIPRR13 pKa = 11.84NDD15 pKa = 3.34TYY17 pKa = 11.48RR18 pKa = 11.84EE19 pKa = 4.08GNRR22 pKa = 11.84IFAWFRR28 pKa = 11.84KK29 pKa = 9.34NDD31 pKa = 2.9IHH33 pKa = 7.66KK34 pKa = 9.66WVLGAEE40 pKa = 4.24KK41 pKa = 10.49GAGGYY46 pKa = 7.24EE47 pKa = 3.88HH48 pKa = 6.81WQMRR52 pKa = 11.84VQCRR56 pKa = 11.84FGFEE60 pKa = 3.86KK61 pKa = 10.89LKK63 pKa = 11.0VLFPNAHH70 pKa = 6.49IEE72 pKa = 4.12EE73 pKa = 5.17CSDD76 pKa = 2.26KK77 pKa = 9.3WTYY80 pKa = 6.97EE81 pKa = 4.19TKK83 pKa = 10.67EE84 pKa = 3.83GVYY87 pKa = 9.18WKK89 pKa = 11.14SNDD92 pKa = 3.33RR93 pKa = 11.84EE94 pKa = 4.2EE95 pKa = 4.28NRR97 pKa = 11.84KK98 pKa = 8.65QRR100 pKa = 11.84FGKK103 pKa = 10.02LRR105 pKa = 11.84YY106 pKa = 7.83AQKK109 pKa = 10.56RR110 pKa = 11.84VIEE113 pKa = 4.52HH114 pKa = 7.36ADD116 pKa = 3.33GTNDD120 pKa = 3.42RR121 pKa = 11.84EE122 pKa = 4.38VVVWYY127 pKa = 9.72DD128 pKa = 3.37QEE130 pKa = 4.44GKK132 pKa = 10.31AGKK135 pKa = 9.48SWLTGAMWEE144 pKa = 4.12RR145 pKa = 11.84GLAYY149 pKa = 9.93FAVADD154 pKa = 4.1TSGKK158 pKa = 8.4TIVMDD163 pKa = 4.01IASEE167 pKa = 4.03YY168 pKa = 10.46LKK170 pKa = 11.03NGWRR174 pKa = 11.84PYY176 pKa = 10.68VIIDD180 pKa = 3.52IPRR183 pKa = 11.84AGKK186 pKa = 7.17WTVIVMTNTMPKK198 pKa = 9.79LDD200 pKa = 4.46KK201 pKa = 11.01LSADD205 pKa = 2.96RR206 pKa = 11.84WKK208 pKa = 10.62IITQEE213 pKa = 3.96EE214 pKa = 4.52LTLEE218 pKa = 4.07EE219 pKa = 3.89TAIEE223 pKa = 4.08QFALTGKK230 pKa = 10.13GGYY233 pKa = 9.12RR234 pKa = 11.84PPLTPPTT241 pKa = 3.94
MM1 pKa = 7.37IMSKK5 pKa = 8.69TFMLTIPRR13 pKa = 11.84NDD15 pKa = 3.34TYY17 pKa = 11.48RR18 pKa = 11.84EE19 pKa = 4.08GNRR22 pKa = 11.84IFAWFRR28 pKa = 11.84KK29 pKa = 9.34NDD31 pKa = 2.9IHH33 pKa = 7.66KK34 pKa = 9.66WVLGAEE40 pKa = 4.24KK41 pKa = 10.49GAGGYY46 pKa = 7.24EE47 pKa = 3.88HH48 pKa = 6.81WQMRR52 pKa = 11.84VQCRR56 pKa = 11.84FGFEE60 pKa = 3.86KK61 pKa = 10.89LKK63 pKa = 11.0VLFPNAHH70 pKa = 6.49IEE72 pKa = 4.12EE73 pKa = 5.17CSDD76 pKa = 2.26KK77 pKa = 9.3WTYY80 pKa = 6.97EE81 pKa = 4.19TKK83 pKa = 10.67EE84 pKa = 3.83GVYY87 pKa = 9.18WKK89 pKa = 11.14SNDD92 pKa = 3.33RR93 pKa = 11.84EE94 pKa = 4.2EE95 pKa = 4.28NRR97 pKa = 11.84KK98 pKa = 8.65QRR100 pKa = 11.84FGKK103 pKa = 10.02LRR105 pKa = 11.84YY106 pKa = 7.83AQKK109 pKa = 10.56RR110 pKa = 11.84VIEE113 pKa = 4.52HH114 pKa = 7.36ADD116 pKa = 3.33GTNDD120 pKa = 3.42RR121 pKa = 11.84EE122 pKa = 4.38VVVWYY127 pKa = 9.72DD128 pKa = 3.37QEE130 pKa = 4.44GKK132 pKa = 10.31AGKK135 pKa = 9.48SWLTGAMWEE144 pKa = 4.12RR145 pKa = 11.84GLAYY149 pKa = 9.93FAVADD154 pKa = 4.1TSGKK158 pKa = 8.4TIVMDD163 pKa = 4.01IASEE167 pKa = 4.03YY168 pKa = 10.46LKK170 pKa = 11.03NGWRR174 pKa = 11.84PYY176 pKa = 10.68VIIDD180 pKa = 3.52IPRR183 pKa = 11.84AGKK186 pKa = 7.17WTVIVMTNTMPKK198 pKa = 9.79LDD200 pKa = 4.46KK201 pKa = 11.01LSADD205 pKa = 2.96RR206 pKa = 11.84WKK208 pKa = 10.62IITQEE213 pKa = 3.96EE214 pKa = 4.52LTLEE218 pKa = 4.07EE219 pKa = 3.89TAIEE223 pKa = 4.08QFALTGKK230 pKa = 10.13GGYY233 pKa = 9.12RR234 pKa = 11.84PPLTPPTT241 pKa = 3.94
Molecular weight: 28.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
658 |
241 |
417 |
329.0 |
36.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.447 ± 0.45 | 1.064 ± 0.13 |
5.623 ± 0.359 | 4.863 ± 1.914 |
3.951 ± 0.121 | 8.967 ± 0.603 |
1.672 ± 0.007 | 6.535 ± 0.173 |
5.471 ± 1.807 | 6.687 ± 0.489 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.191 ± 0.071 | 4.559 ± 0.46 |
5.167 ± 0.798 | 3.191 ± 0.16 |
5.319 ± 0.967 | 6.839 ± 2.192 |
7.903 ± 0.242 | 5.471 ± 0.043 |
2.432 ± 1.188 | 3.647 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
