
Methylobacterium tarhaniae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
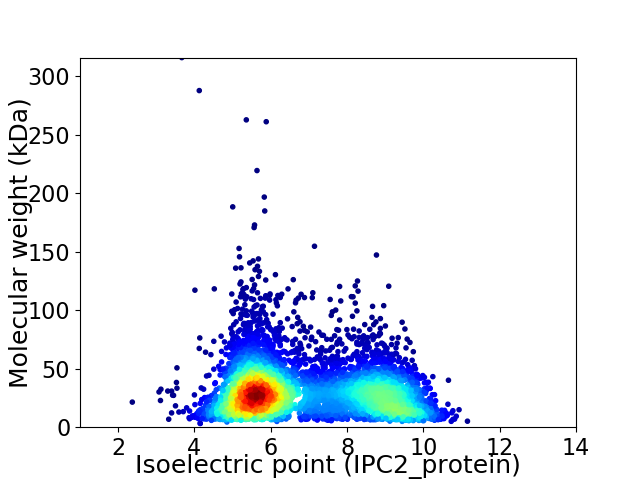
Virtual 2D-PAGE plot for 5746 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J6V5T0|A0A0J6V5T0_9RHIZ Beta-ketoacyl synthase OS=Methylobacterium tarhaniae OX=1187852 GN=VQ03_23540 PE=3 SV=1
MM1 pKa = 7.13ATILGTNGNDD11 pKa = 3.04VLTGTTGDD19 pKa = 3.9DD20 pKa = 3.92VILGLLGNDD29 pKa = 4.31RR30 pKa = 11.84ISDD33 pKa = 3.46PGGFNRR39 pKa = 11.84IDD41 pKa = 3.75GQDD44 pKa = 3.5GADD47 pKa = 3.92VITGGANLDD56 pKa = 4.32YY57 pKa = 10.87IAGGPGNDD65 pKa = 3.36VIYY68 pKa = 10.63GGGGADD74 pKa = 3.55QLIGEE79 pKa = 5.02AGDD82 pKa = 3.81DD83 pKa = 4.54LIYY86 pKa = 10.86GQDD89 pKa = 3.55GDD91 pKa = 5.55DD92 pKa = 3.75YY93 pKa = 11.91AAGNPGNDD101 pKa = 3.74TIYY104 pKa = 11.07GGAGNDD110 pKa = 3.53FFVGEE115 pKa = 4.06QGNDD119 pKa = 3.18QVYY122 pKa = 10.66GEE124 pKa = 4.77AGNDD128 pKa = 3.61FVAGGEE134 pKa = 4.03DD135 pKa = 3.98DD136 pKa = 5.35DD137 pKa = 5.28LVSGGDD143 pKa = 3.64GDD145 pKa = 5.31DD146 pKa = 5.34LVDD149 pKa = 4.57GDD151 pKa = 5.14LGNDD155 pKa = 3.66TLLGDD160 pKa = 4.6AGNDD164 pKa = 3.46VLFGDD169 pKa = 4.34YY170 pKa = 11.29GNDD173 pKa = 3.29RR174 pKa = 11.84MNGGPGNDD182 pKa = 3.47RR183 pKa = 11.84LDD185 pKa = 3.91GAVGTDD191 pKa = 3.04TAVFDD196 pKa = 3.67TAFRR200 pKa = 11.84NLRR203 pKa = 11.84VTSSGSLVTFEE214 pKa = 4.39GATGIDD220 pKa = 3.42EE221 pKa = 4.54VKK223 pKa = 9.32NTEE226 pKa = 3.76VFEE229 pKa = 4.37FSDD232 pKa = 3.64RR233 pKa = 11.84TIVQADD239 pKa = 3.44GNAAVDD245 pKa = 3.61DD246 pKa = 4.81LYY248 pKa = 11.65YY249 pKa = 10.88LSRR252 pKa = 11.84NADD255 pKa = 3.38VLLAGLDD262 pKa = 3.73AEE264 pKa = 4.61AHH266 pKa = 6.03FGQYY270 pKa = 7.65GWRR273 pKa = 11.84EE274 pKa = 3.68GRR276 pKa = 11.84NPNAYY281 pKa = 9.96FDD283 pKa = 3.99TKK285 pKa = 10.9GYY287 pKa = 9.62LAAYY291 pKa = 9.62SDD293 pKa = 3.9VAAAGIDD300 pKa = 3.76PLQHH304 pKa = 5.55YY305 pKa = 8.23LQYY308 pKa = 10.55GWKK311 pKa = 9.81EE312 pKa = 3.86GRR314 pKa = 11.84DD315 pKa = 3.41PSANFDD321 pKa = 3.57TKK323 pKa = 11.13AYY325 pKa = 10.23LAANPDD331 pKa = 3.2VAAAGINPLEE341 pKa = 4.8HH342 pKa = 6.45FLQYY346 pKa = 11.23GSVEE350 pKa = 3.99GRR352 pKa = 11.84AVQPGDD358 pKa = 3.38GAFATATAPGVYY370 pKa = 8.56TT371 pKa = 4.26
MM1 pKa = 7.13ATILGTNGNDD11 pKa = 3.04VLTGTTGDD19 pKa = 3.9DD20 pKa = 3.92VILGLLGNDD29 pKa = 4.31RR30 pKa = 11.84ISDD33 pKa = 3.46PGGFNRR39 pKa = 11.84IDD41 pKa = 3.75GQDD44 pKa = 3.5GADD47 pKa = 3.92VITGGANLDD56 pKa = 4.32YY57 pKa = 10.87IAGGPGNDD65 pKa = 3.36VIYY68 pKa = 10.63GGGGADD74 pKa = 3.55QLIGEE79 pKa = 5.02AGDD82 pKa = 3.81DD83 pKa = 4.54LIYY86 pKa = 10.86GQDD89 pKa = 3.55GDD91 pKa = 5.55DD92 pKa = 3.75YY93 pKa = 11.91AAGNPGNDD101 pKa = 3.74TIYY104 pKa = 11.07GGAGNDD110 pKa = 3.53FFVGEE115 pKa = 4.06QGNDD119 pKa = 3.18QVYY122 pKa = 10.66GEE124 pKa = 4.77AGNDD128 pKa = 3.61FVAGGEE134 pKa = 4.03DD135 pKa = 3.98DD136 pKa = 5.35DD137 pKa = 5.28LVSGGDD143 pKa = 3.64GDD145 pKa = 5.31DD146 pKa = 5.34LVDD149 pKa = 4.57GDD151 pKa = 5.14LGNDD155 pKa = 3.66TLLGDD160 pKa = 4.6AGNDD164 pKa = 3.46VLFGDD169 pKa = 4.34YY170 pKa = 11.29GNDD173 pKa = 3.29RR174 pKa = 11.84MNGGPGNDD182 pKa = 3.47RR183 pKa = 11.84LDD185 pKa = 3.91GAVGTDD191 pKa = 3.04TAVFDD196 pKa = 3.67TAFRR200 pKa = 11.84NLRR203 pKa = 11.84VTSSGSLVTFEE214 pKa = 4.39GATGIDD220 pKa = 3.42EE221 pKa = 4.54VKK223 pKa = 9.32NTEE226 pKa = 3.76VFEE229 pKa = 4.37FSDD232 pKa = 3.64RR233 pKa = 11.84TIVQADD239 pKa = 3.44GNAAVDD245 pKa = 3.61DD246 pKa = 4.81LYY248 pKa = 11.65YY249 pKa = 10.88LSRR252 pKa = 11.84NADD255 pKa = 3.38VLLAGLDD262 pKa = 3.73AEE264 pKa = 4.61AHH266 pKa = 6.03FGQYY270 pKa = 7.65GWRR273 pKa = 11.84EE274 pKa = 3.68GRR276 pKa = 11.84NPNAYY281 pKa = 9.96FDD283 pKa = 3.99TKK285 pKa = 10.9GYY287 pKa = 9.62LAAYY291 pKa = 9.62SDD293 pKa = 3.9VAAAGIDD300 pKa = 3.76PLQHH304 pKa = 5.55YY305 pKa = 8.23LQYY308 pKa = 10.55GWKK311 pKa = 9.81EE312 pKa = 3.86GRR314 pKa = 11.84DD315 pKa = 3.41PSANFDD321 pKa = 3.57TKK323 pKa = 11.13AYY325 pKa = 10.23LAANPDD331 pKa = 3.2VAAAGINPLEE341 pKa = 4.8HH342 pKa = 6.45FLQYY346 pKa = 11.23GSVEE350 pKa = 3.99GRR352 pKa = 11.84AVQPGDD358 pKa = 3.38GAFATATAPGVYY370 pKa = 8.56TT371 pKa = 4.26
Molecular weight: 38.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J6SXX4|A0A0J6SXX4_9RHIZ Uncharacterized protein OS=Methylobacterium tarhaniae OX=1187852 GN=VQ03_16955 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.31VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1734764 |
29 |
3241 |
301.9 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.485 ± 0.055 | 0.808 ± 0.01 |
5.471 ± 0.026 | 5.504 ± 0.03 |
3.296 ± 0.022 | 9.257 ± 0.038 |
1.951 ± 0.016 | 4.2 ± 0.027 |
2.325 ± 0.024 | 10.679 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.05 ± 0.015 | 1.962 ± 0.02 |
5.946 ± 0.036 | 2.772 ± 0.019 |
8.282 ± 0.038 | 4.704 ± 0.023 |
5.271 ± 0.029 | 7.76 ± 0.031 |
1.259 ± 0.012 | 2.019 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
