
Chicken associated cyclovirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus
Average proteome isoelectric point is 8.88
Get precalculated fractions of proteins
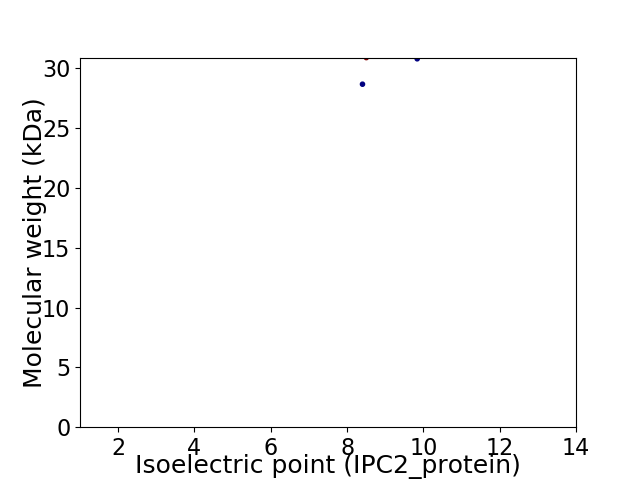
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BLF6|A0A346BLF6_9CIRC Capsid protein OS=Chicken associated cyclovirus 2 OX=2109365 PE=4 SV=1
MM1 pKa = 7.25LRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84FGSIKK11 pKa = 10.25KK12 pKa = 9.55ILGHH16 pKa = 6.89RR17 pKa = 11.84IHH19 pKa = 7.42IEE21 pKa = 3.7RR22 pKa = 11.84SNGSDD27 pKa = 2.9VDD29 pKa = 3.78NQKK32 pKa = 10.39YY33 pKa = 9.66CSKK36 pKa = 11.17GGDD39 pKa = 3.49FWEE42 pKa = 5.04HH43 pKa = 6.86GSPQGQGKK51 pKa = 10.26RR52 pKa = 11.84NDD54 pKa = 4.18LISIVEE60 pKa = 4.4EE61 pKa = 4.09IKK63 pKa = 11.03SGTGLVEE70 pKa = 3.35ICEE73 pKa = 4.12NHH75 pKa = 5.68TEE77 pKa = 4.11SFIRR81 pKa = 11.84YY82 pKa = 8.64HH83 pKa = 7.82SGIEE87 pKa = 3.55KK88 pKa = 9.65AYY90 pKa = 9.66RR91 pKa = 11.84YY92 pKa = 9.45IGRR95 pKa = 11.84GCNDD99 pKa = 3.8RR100 pKa = 11.84NWKK103 pKa = 8.5TKK105 pKa = 10.06VFTPQTSWGASLMFFLHH122 pKa = 5.87VFYY125 pKa = 10.72GATGTGKK132 pKa = 10.25SRR134 pKa = 11.84LAALRR139 pKa = 11.84CSGRR143 pKa = 11.84RR144 pKa = 11.84VYY146 pKa = 10.52YY147 pKa = 10.24KK148 pKa = 10.53PHH150 pKa = 6.69DD151 pKa = 4.02KK152 pKa = 9.93WWDD155 pKa = 3.69GYY157 pKa = 11.08DD158 pKa = 3.2GHH160 pKa = 6.74EE161 pKa = 4.62CVIIDD166 pKa = 5.35DD167 pKa = 4.65FYY169 pKa = 11.58GWIPYY174 pKa = 10.23DD175 pKa = 3.5EE176 pKa = 5.12LLRR179 pKa = 11.84ITDD182 pKa = 4.37RR183 pKa = 11.84YY184 pKa = 7.42PHH186 pKa = 6.39RR187 pKa = 11.84VAIKK191 pKa = 9.53GAMIQFLAKK200 pKa = 10.3DD201 pKa = 3.61IYY203 pKa = 9.76ITSNKK208 pKa = 9.19HH209 pKa = 4.48PRR211 pKa = 11.84DD212 pKa = 3.62WYY214 pKa = 9.09NHH216 pKa = 5.8LWFQDD221 pKa = 3.36NQYY224 pKa = 11.16AALKK228 pKa = 10.32RR229 pKa = 11.84RR230 pKa = 11.84IDD232 pKa = 3.28EE233 pKa = 4.68HH234 pKa = 7.85ICMGEE239 pKa = 4.04GFNKK243 pKa = 10.11EE244 pKa = 3.89LL245 pKa = 4.27
MM1 pKa = 7.25LRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84FGSIKK11 pKa = 10.25KK12 pKa = 9.55ILGHH16 pKa = 6.89RR17 pKa = 11.84IHH19 pKa = 7.42IEE21 pKa = 3.7RR22 pKa = 11.84SNGSDD27 pKa = 2.9VDD29 pKa = 3.78NQKK32 pKa = 10.39YY33 pKa = 9.66CSKK36 pKa = 11.17GGDD39 pKa = 3.49FWEE42 pKa = 5.04HH43 pKa = 6.86GSPQGQGKK51 pKa = 10.26RR52 pKa = 11.84NDD54 pKa = 4.18LISIVEE60 pKa = 4.4EE61 pKa = 4.09IKK63 pKa = 11.03SGTGLVEE70 pKa = 3.35ICEE73 pKa = 4.12NHH75 pKa = 5.68TEE77 pKa = 4.11SFIRR81 pKa = 11.84YY82 pKa = 8.64HH83 pKa = 7.82SGIEE87 pKa = 3.55KK88 pKa = 9.65AYY90 pKa = 9.66RR91 pKa = 11.84YY92 pKa = 9.45IGRR95 pKa = 11.84GCNDD99 pKa = 3.8RR100 pKa = 11.84NWKK103 pKa = 8.5TKK105 pKa = 10.06VFTPQTSWGASLMFFLHH122 pKa = 5.87VFYY125 pKa = 10.72GATGTGKK132 pKa = 10.25SRR134 pKa = 11.84LAALRR139 pKa = 11.84CSGRR143 pKa = 11.84RR144 pKa = 11.84VYY146 pKa = 10.52YY147 pKa = 10.24KK148 pKa = 10.53PHH150 pKa = 6.69DD151 pKa = 4.02KK152 pKa = 9.93WWDD155 pKa = 3.69GYY157 pKa = 11.08DD158 pKa = 3.2GHH160 pKa = 6.74EE161 pKa = 4.62CVIIDD166 pKa = 5.35DD167 pKa = 4.65FYY169 pKa = 11.58GWIPYY174 pKa = 10.23DD175 pKa = 3.5EE176 pKa = 5.12LLRR179 pKa = 11.84ITDD182 pKa = 4.37RR183 pKa = 11.84YY184 pKa = 7.42PHH186 pKa = 6.39RR187 pKa = 11.84VAIKK191 pKa = 9.53GAMIQFLAKK200 pKa = 10.3DD201 pKa = 3.61IYY203 pKa = 9.76ITSNKK208 pKa = 9.19HH209 pKa = 4.48PRR211 pKa = 11.84DD212 pKa = 3.62WYY214 pKa = 9.09NHH216 pKa = 5.8LWFQDD221 pKa = 3.36NQYY224 pKa = 11.16AALKK228 pKa = 10.32RR229 pKa = 11.84RR230 pKa = 11.84IDD232 pKa = 3.28EE233 pKa = 4.68HH234 pKa = 7.85ICMGEE239 pKa = 4.04GFNKK243 pKa = 10.11EE244 pKa = 3.89LL245 pKa = 4.27
Molecular weight: 28.68 kDa
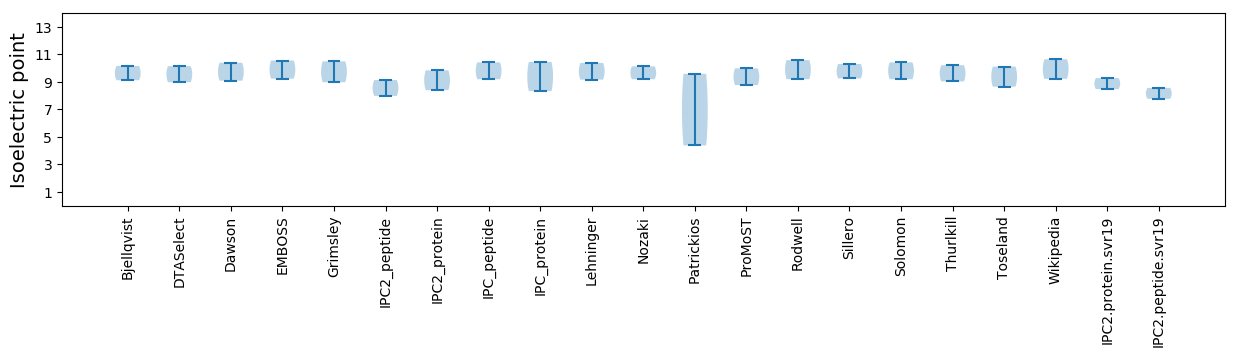
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BLF6|A0A346BLF6_9CIRC Capsid protein OS=Chicken associated cyclovirus 2 OX=2109365 PE=4 SV=1
MM1 pKa = 7.59AFFPRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LFRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84YY17 pKa = 8.43NLKK20 pKa = 9.72RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 8.59FRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84FMRR32 pKa = 11.84SSRR35 pKa = 11.84ATTRR39 pKa = 11.84HH40 pKa = 4.92EE41 pKa = 4.03LSRR44 pKa = 11.84GGQLFTRR51 pKa = 11.84LHH53 pKa = 5.34QSYY56 pKa = 10.16ILNSTTGRR64 pKa = 11.84TIYY67 pKa = 10.23LAPQLQSFTHH77 pKa = 5.0YY78 pKa = 10.26TNFAQFWSSIRR89 pKa = 11.84VWKK92 pKa = 9.92IAYY95 pKa = 8.9KK96 pKa = 7.88ITPVITEE103 pKa = 4.29NVPIDD108 pKa = 3.84NAGATYY114 pKa = 9.99VLSFGRR120 pKa = 11.84HH121 pKa = 4.55VGSIYY126 pKa = 10.78YY127 pKa = 10.14DD128 pKa = 3.51DD129 pKa = 4.41QFPGAANTGVAQSYY143 pKa = 10.36YY144 pKa = 11.0DD145 pKa = 4.23EE146 pKa = 4.93ISNFPNSKK154 pKa = 8.59STSITKK160 pKa = 9.39PLRR163 pKa = 11.84FSFVPKK169 pKa = 9.52TVVNAVGVNGTITGKK184 pKa = 10.4GWVARR189 pKa = 11.84RR190 pKa = 11.84KK191 pKa = 9.67LDD193 pKa = 3.52CDD195 pKa = 3.68TSVGTSLYY203 pKa = 10.52GLVYY207 pKa = 9.99CYY209 pKa = 10.04EE210 pKa = 4.29GQQPEE215 pKa = 4.33STNIQVRR222 pKa = 11.84VDD224 pKa = 3.36CYY226 pKa = 10.68IYY228 pKa = 10.17ATFYY232 pKa = 10.64EE233 pKa = 4.47YY234 pKa = 10.77RR235 pKa = 11.84QFQYY239 pKa = 11.22SPTLEE244 pKa = 4.89KK245 pKa = 9.99IDD247 pKa = 4.47LNKK250 pKa = 9.93MDD252 pKa = 4.76INKK255 pKa = 9.36IDD257 pKa = 3.85DD258 pKa = 4.08KK259 pKa = 10.74KK260 pKa = 9.68TFMM263 pKa = 5.4
MM1 pKa = 7.59AFFPRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84LFRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PRR16 pKa = 11.84YY17 pKa = 8.43NLKK20 pKa = 9.72RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 8.59FRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84FMRR32 pKa = 11.84SSRR35 pKa = 11.84ATTRR39 pKa = 11.84HH40 pKa = 4.92EE41 pKa = 4.03LSRR44 pKa = 11.84GGQLFTRR51 pKa = 11.84LHH53 pKa = 5.34QSYY56 pKa = 10.16ILNSTTGRR64 pKa = 11.84TIYY67 pKa = 10.23LAPQLQSFTHH77 pKa = 5.0YY78 pKa = 10.26TNFAQFWSSIRR89 pKa = 11.84VWKK92 pKa = 9.92IAYY95 pKa = 8.9KK96 pKa = 7.88ITPVITEE103 pKa = 4.29NVPIDD108 pKa = 3.84NAGATYY114 pKa = 9.99VLSFGRR120 pKa = 11.84HH121 pKa = 4.55VGSIYY126 pKa = 10.78YY127 pKa = 10.14DD128 pKa = 3.51DD129 pKa = 4.41QFPGAANTGVAQSYY143 pKa = 10.36YY144 pKa = 11.0DD145 pKa = 4.23EE146 pKa = 4.93ISNFPNSKK154 pKa = 8.59STSITKK160 pKa = 9.39PLRR163 pKa = 11.84FSFVPKK169 pKa = 9.52TVVNAVGVNGTITGKK184 pKa = 10.4GWVARR189 pKa = 11.84RR190 pKa = 11.84KK191 pKa = 9.67LDD193 pKa = 3.52CDD195 pKa = 3.68TSVGTSLYY203 pKa = 10.52GLVYY207 pKa = 9.99CYY209 pKa = 10.04EE210 pKa = 4.29GQQPEE215 pKa = 4.33STNIQVRR222 pKa = 11.84VDD224 pKa = 3.36CYY226 pKa = 10.68IYY228 pKa = 10.17ATFYY232 pKa = 10.64EE233 pKa = 4.47YY234 pKa = 10.77RR235 pKa = 11.84QFQYY239 pKa = 11.22SPTLEE244 pKa = 4.89KK245 pKa = 9.99IDD247 pKa = 4.47LNKK250 pKa = 9.93MDD252 pKa = 4.76INKK255 pKa = 9.36IDD257 pKa = 3.85DD258 pKa = 4.08KK259 pKa = 10.74KK260 pKa = 9.68TFMM263 pKa = 5.4
Molecular weight: 30.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
508 |
245 |
263 |
254.0 |
29.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.724 ± 0.148 | 1.772 ± 0.428 |
5.315 ± 0.768 | 3.937 ± 0.865 |
5.512 ± 0.646 | 7.677 ± 1.339 |
3.15 ± 1.105 | 7.283 ± 0.814 |
5.906 ± 0.653 | 5.709 ± 0.004 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.575 ± 0.037 | 4.528 ± 0.282 |
3.346 ± 0.567 | 3.74 ± 0.558 |
9.449 ± 0.813 | 6.693 ± 0.619 |
6.299 ± 1.66 | 4.724 ± 0.922 |
2.165 ± 0.695 | 6.496 ± 0.494 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
