
African cassava mosaic virus (isolate West Kenyan 844) (ACMV) (Cassava latent virus (isolate West Kenyan 844))
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; African cassava mosaic virus
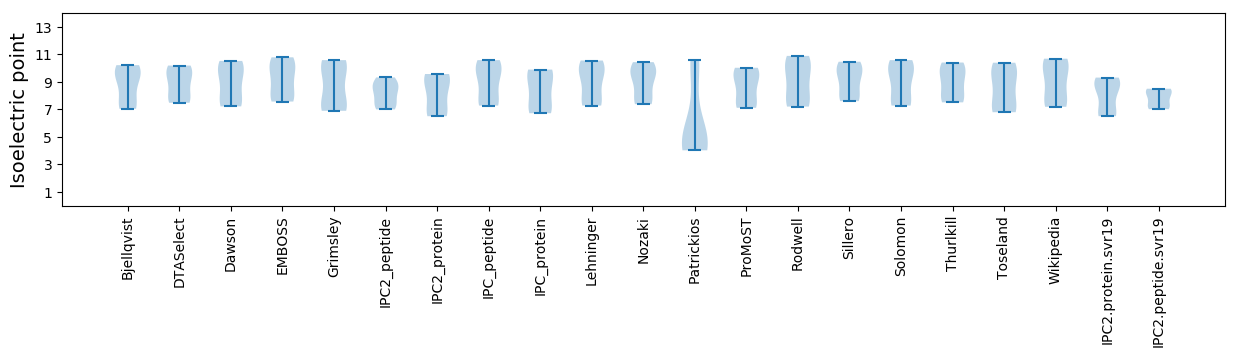
Average proteome isoelectric point is 8.05
Get precalculated fractions of proteins
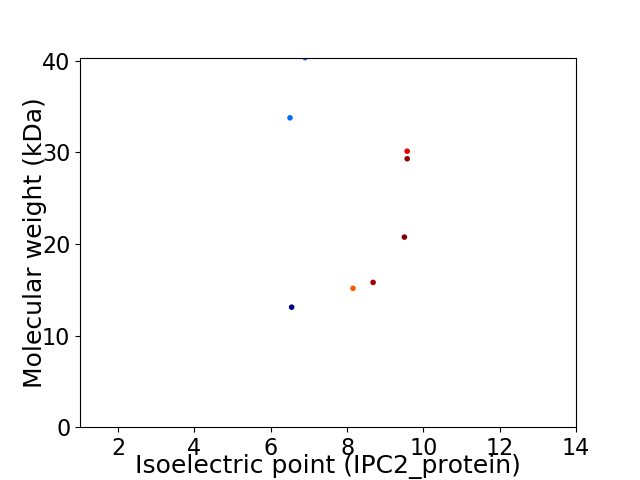
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P14982|REP_CLVK Replication-associated protein OS=African cassava mosaic virus (isolate West Kenyan 844) OX=10818 GN=AC1 PE=3 SV=1
MM1 pKa = 7.47DD2 pKa = 3.97TSVPVISSDD11 pKa = 3.91YY12 pKa = 8.59IHH14 pKa = 7.09SARR17 pKa = 11.84TEE19 pKa = 3.99YY20 pKa = 11.33KK21 pKa = 9.37LTNDD25 pKa = 3.85EE26 pKa = 5.06SPITLQFPSTLEE38 pKa = 3.82RR39 pKa = 11.84TRR41 pKa = 11.84VRR43 pKa = 11.84IMGKK47 pKa = 9.13CMKK50 pKa = 9.42VDD52 pKa = 3.32HH53 pKa = 6.42VVIEE57 pKa = 4.18YY58 pKa = 10.56RR59 pKa = 11.84NQVPFNAQGSVIVTIRR75 pKa = 11.84DD76 pKa = 3.29TRR78 pKa = 11.84LSDD81 pKa = 3.52EE82 pKa = 4.56QQDD85 pKa = 3.6QAQFTFPIGCNVDD98 pKa = 2.92LHH100 pKa = 6.37YY101 pKa = 10.89FSASYY106 pKa = 9.98FSIDD110 pKa = 5.73DD111 pKa = 3.72NVPWQLLYY119 pKa = 10.6KK120 pKa = 10.56VEE122 pKa = 4.46DD123 pKa = 3.88SNVKK127 pKa = 10.56NGITFAQIKK136 pKa = 10.36AKK138 pKa = 10.61LKK140 pKa = 10.61LSAAKK145 pKa = 10.11HH146 pKa = 4.81STDD149 pKa = 3.87IKK151 pKa = 10.67FKK153 pKa = 10.86QPTIKK158 pKa = 10.14ILSKK162 pKa = 10.75DD163 pKa = 3.81YY164 pKa = 11.52GPDD167 pKa = 3.6CVDD170 pKa = 4.09FWSVGKK176 pKa = 9.26PKK178 pKa = 10.29PIRR181 pKa = 11.84RR182 pKa = 11.84LIQNEE187 pKa = 4.1PGTDD191 pKa = 3.36YY192 pKa = 10.14DD193 pKa = 3.64TGPKK197 pKa = 9.31YY198 pKa = 10.55RR199 pKa = 11.84PITVQPGEE207 pKa = 3.88TWATKK212 pKa = 8.57STIGRR217 pKa = 11.84YY218 pKa = 7.16TSMRR222 pKa = 11.84YY223 pKa = 7.26TRR225 pKa = 11.84PNPIDD230 pKa = 3.84IDD232 pKa = 4.1DD233 pKa = 4.53SSSKK237 pKa = 10.61QYY239 pKa = 10.31TSEE242 pKa = 4.31AEE244 pKa = 3.91FPLRR248 pKa = 11.84GLHH251 pKa = 5.61QLPEE255 pKa = 4.2ASLDD259 pKa = 3.78PGDD262 pKa = 5.63SISQTQSMSKK272 pKa = 10.41KK273 pKa = 9.69DD274 pKa = 3.24IEE276 pKa = 4.67SIIEE280 pKa = 3.86QTVNKK285 pKa = 9.95CLIAHH290 pKa = 7.22RR291 pKa = 11.84GSSHH295 pKa = 7.39KK296 pKa = 10.74DD297 pKa = 3.0LL298 pKa = 5.31
MM1 pKa = 7.47DD2 pKa = 3.97TSVPVISSDD11 pKa = 3.91YY12 pKa = 8.59IHH14 pKa = 7.09SARR17 pKa = 11.84TEE19 pKa = 3.99YY20 pKa = 11.33KK21 pKa = 9.37LTNDD25 pKa = 3.85EE26 pKa = 5.06SPITLQFPSTLEE38 pKa = 3.82RR39 pKa = 11.84TRR41 pKa = 11.84VRR43 pKa = 11.84IMGKK47 pKa = 9.13CMKK50 pKa = 9.42VDD52 pKa = 3.32HH53 pKa = 6.42VVIEE57 pKa = 4.18YY58 pKa = 10.56RR59 pKa = 11.84NQVPFNAQGSVIVTIRR75 pKa = 11.84DD76 pKa = 3.29TRR78 pKa = 11.84LSDD81 pKa = 3.52EE82 pKa = 4.56QQDD85 pKa = 3.6QAQFTFPIGCNVDD98 pKa = 2.92LHH100 pKa = 6.37YY101 pKa = 10.89FSASYY106 pKa = 9.98FSIDD110 pKa = 5.73DD111 pKa = 3.72NVPWQLLYY119 pKa = 10.6KK120 pKa = 10.56VEE122 pKa = 4.46DD123 pKa = 3.88SNVKK127 pKa = 10.56NGITFAQIKK136 pKa = 10.36AKK138 pKa = 10.61LKK140 pKa = 10.61LSAAKK145 pKa = 10.11HH146 pKa = 4.81STDD149 pKa = 3.87IKK151 pKa = 10.67FKK153 pKa = 10.86QPTIKK158 pKa = 10.14ILSKK162 pKa = 10.75DD163 pKa = 3.81YY164 pKa = 11.52GPDD167 pKa = 3.6CVDD170 pKa = 4.09FWSVGKK176 pKa = 9.26PKK178 pKa = 10.29PIRR181 pKa = 11.84RR182 pKa = 11.84LIQNEE187 pKa = 4.1PGTDD191 pKa = 3.36YY192 pKa = 10.14DD193 pKa = 3.64TGPKK197 pKa = 9.31YY198 pKa = 10.55RR199 pKa = 11.84PITVQPGEE207 pKa = 3.88TWATKK212 pKa = 8.57STIGRR217 pKa = 11.84YY218 pKa = 7.16TSMRR222 pKa = 11.84YY223 pKa = 7.26TRR225 pKa = 11.84PNPIDD230 pKa = 3.84IDD232 pKa = 4.1DD233 pKa = 4.53SSSKK237 pKa = 10.61QYY239 pKa = 10.31TSEE242 pKa = 4.31AEE244 pKa = 3.91FPLRR248 pKa = 11.84GLHH251 pKa = 5.61QLPEE255 pKa = 4.2ASLDD259 pKa = 3.78PGDD262 pKa = 5.63SISQTQSMSKK272 pKa = 10.41KK273 pKa = 9.69DD274 pKa = 3.24IEE276 pKa = 4.67SIIEE280 pKa = 3.86QTVNKK285 pKa = 9.95CLIAHH290 pKa = 7.22RR291 pKa = 11.84GSSHH295 pKa = 7.39KK296 pKa = 10.74DD297 pKa = 3.0LL298 pKa = 5.31
Molecular weight: 33.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P14975|AV2_CLVK Protein AV2 OS=African cassava mosaic virus (isolate West Kenyan 844) OX=10818 PE=3 SV=1
MM1 pKa = 8.05PFRR4 pKa = 11.84DD5 pKa = 4.21TYY7 pKa = 11.02IMSPINRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84CSRR19 pKa = 11.84VSLVEE24 pKa = 3.91CLQLTWSTXEE34 pKa = 4.15LLVFEE39 pKa = 5.45FKK41 pKa = 10.66PRR43 pKa = 11.84MSFSHH48 pKa = 5.46TQSVLYY54 pKa = 9.45PKK56 pKa = 9.15NTYY59 pKa = 9.51CHH61 pKa = 6.02SFKK64 pKa = 10.88HH65 pKa = 5.53FLSNQTLSSLKK76 pKa = 10.5SVEE79 pKa = 3.91SCIRR83 pKa = 11.84MGNLTCMPSFNSRR96 pKa = 11.84VKK98 pKa = 10.55SRR100 pKa = 11.84LRR102 pKa = 11.84TIVSSIVYY110 pKa = 7.12TQAVAPVSTPTFKK123 pKa = 11.01VPNQARR129 pKa = 11.84MSSPIWIRR137 pKa = 11.84TATPSNGDD145 pKa = 3.33NFKK148 pKa = 11.51SMDD151 pKa = 4.11DD152 pKa = 3.52LLEE155 pKa = 4.22AVNNQRR161 pKa = 11.84MMLTPKK167 pKa = 10.4RR168 pKa = 11.84LTAAVSQKK176 pKa = 11.4LLMSLGNN183 pKa = 3.61
MM1 pKa = 8.05PFRR4 pKa = 11.84DD5 pKa = 4.21TYY7 pKa = 11.02IMSPINRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84CSRR19 pKa = 11.84VSLVEE24 pKa = 3.91CLQLTWSTXEE34 pKa = 4.15LLVFEE39 pKa = 5.45FKK41 pKa = 10.66PRR43 pKa = 11.84MSFSHH48 pKa = 5.46TQSVLYY54 pKa = 9.45PKK56 pKa = 9.15NTYY59 pKa = 9.51CHH61 pKa = 6.02SFKK64 pKa = 10.88HH65 pKa = 5.53FLSNQTLSSLKK76 pKa = 10.5SVEE79 pKa = 3.91SCIRR83 pKa = 11.84MGNLTCMPSFNSRR96 pKa = 11.84VKK98 pKa = 10.55SRR100 pKa = 11.84LRR102 pKa = 11.84TIVSSIVYY110 pKa = 7.12TQAVAPVSTPTFKK123 pKa = 11.01VPNQARR129 pKa = 11.84MSSPIWIRR137 pKa = 11.84TATPSNGDD145 pKa = 3.33NFKK148 pKa = 11.51SMDD151 pKa = 4.11DD152 pKa = 3.52LLEE155 pKa = 4.22AVNNQRR161 pKa = 11.84MMLTPKK167 pKa = 10.4RR168 pKa = 11.84LTAAVSQKK176 pKa = 11.4LLMSLGNN183 pKa = 3.61
Molecular weight: 20.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1735 |
113 |
358 |
216.9 |
24.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.611 ± 0.423 | 2.017 ± 0.207 |
4.784 ± 0.631 | 4.035 ± 0.412 |
4.496 ± 0.373 | 5.014 ± 0.483 |
3.804 ± 0.587 | 6.052 ± 0.794 |
5.764 ± 0.452 | 7.205 ± 0.534 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.017 ± 0.448 | 5.821 ± 0.591 |
5.937 ± 0.35 | 5.13 ± 0.398 |
6.744 ± 0.755 | 8.818 ± 1.299 |
5.937 ± 0.581 | 6.571 ± 0.57 |
1.21 ± 0.176 | 3.977 ± 0.505 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
