
Oceanobacillus oncorhynchi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Oceanobacillus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
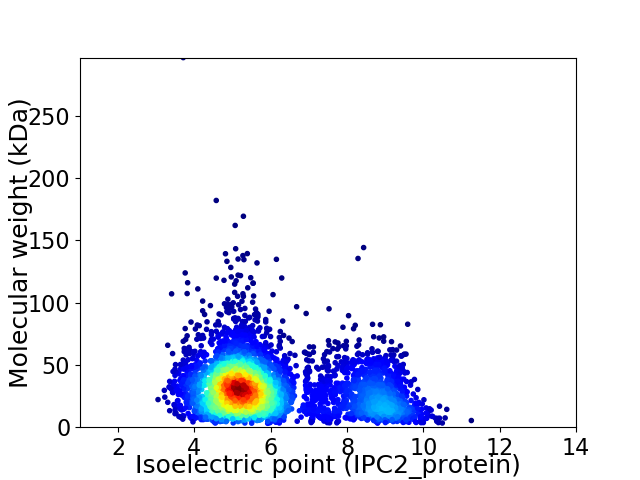
Virtual 2D-PAGE plot for 4386 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A1MLP3|A0A0A1MLP3_9BACI Cystathionine gamma-synthase/O-acetylhomoserine (Thiol)-lyase OS=Oceanobacillus oncorhynchi OX=545501 GN=metI PE=3 SV=1
MM1 pKa = 7.18LKK3 pKa = 9.95KK4 pKa = 9.75WFFFIATLVFVSALYY19 pKa = 10.47ACSGGDD25 pKa = 4.17DD26 pKa = 3.8GDD28 pKa = 4.2SKK30 pKa = 11.1EE31 pKa = 4.22GEE33 pKa = 4.08AAGGDD38 pKa = 3.74NVNATGFPIVEE49 pKa = 4.49EE50 pKa = 4.05EE51 pKa = 4.42LEE53 pKa = 4.1LTIFANKK60 pKa = 8.56PAQNEE65 pKa = 4.15GNDD68 pKa = 3.57WNDD71 pKa = 2.75ILIWNTYY78 pKa = 10.16RR79 pKa = 11.84DD80 pKa = 3.75LTNINVNWNSVSPDD94 pKa = 3.09ALEE97 pKa = 4.15EE98 pKa = 3.98NRR100 pKa = 11.84NLALGSGDD108 pKa = 4.09LPDD111 pKa = 4.49AFFLAEE117 pKa = 4.79LSNTDD122 pKa = 3.68LLRR125 pKa = 11.84YY126 pKa = 9.34GSQGAFLPLNDD137 pKa = 5.84LIDD140 pKa = 4.31EE141 pKa = 4.35YY142 pKa = 11.74APNLKK147 pKa = 10.42ALMEE151 pKa = 4.59NDD153 pKa = 3.42PTIEE157 pKa = 4.04KK158 pKa = 10.55AITFPDD164 pKa = 3.68GNIYY168 pKa = 10.87SMPSLIEE175 pKa = 4.77DD176 pKa = 4.05DD177 pKa = 4.36FLSLRR182 pKa = 11.84LSARR186 pKa = 11.84PWVNEE191 pKa = 3.52DD192 pKa = 3.02WLNEE196 pKa = 3.75LDD198 pKa = 3.64MDD200 pKa = 4.28IPEE203 pKa = 4.45TTGDD207 pKa = 3.31FYY209 pKa = 11.51EE210 pKa = 4.13YY211 pKa = 10.74LKK213 pKa = 10.83AVKK216 pKa = 10.14EE217 pKa = 3.99LDD219 pKa = 3.43PVGNGDD225 pKa = 4.24TIPYY229 pKa = 9.4GGTDD233 pKa = 2.94IAEE236 pKa = 4.22LVQWLSGSFGVMNHH250 pKa = 6.26GASNTNLDD258 pKa = 4.69LDD260 pKa = 4.75PNDD263 pKa = 3.95DD264 pKa = 3.8SKK266 pKa = 11.74VRR268 pKa = 11.84YY269 pKa = 8.93YY270 pKa = 10.46ATADD274 pKa = 3.45EE275 pKa = 4.16YY276 pKa = 11.48RR277 pKa = 11.84EE278 pKa = 3.92MLEE281 pKa = 4.31YY282 pKa = 10.42IHH284 pKa = 6.45MLYY287 pKa = 10.67EE288 pKa = 4.35EE289 pKa = 4.44GLIDD293 pKa = 4.2QSIFTIEE300 pKa = 3.65WGQFLANASDD310 pKa = 3.62NLYY313 pKa = 11.17GSMIFYY319 pKa = 10.71DD320 pKa = 4.5PIEE323 pKa = 4.34LFGEE327 pKa = 4.68EE328 pKa = 4.82IGSQYY333 pKa = 11.39NSLAALEE340 pKa = 5.1GPDD343 pKa = 3.69GHH345 pKa = 6.63QSYY348 pKa = 11.15NKK350 pKa = 9.86LSSSVWTPDD359 pKa = 2.93NLVITSEE366 pKa = 4.25NPNPAATVRR375 pKa = 11.84WMDD378 pKa = 3.36HH379 pKa = 6.28FYY381 pKa = 11.23GDD383 pKa = 3.91EE384 pKa = 4.15GAEE387 pKa = 3.99LYY389 pKa = 11.42YY390 pKa = 10.25MGVEE394 pKa = 4.09GEE396 pKa = 4.28TFEE399 pKa = 5.76IEE401 pKa = 4.17DD402 pKa = 4.0GEE404 pKa = 4.55AVYY407 pKa = 10.79SDD409 pKa = 5.26HH410 pKa = 7.0ILNPEE415 pKa = 3.49GDD417 pKa = 3.72MTFEE421 pKa = 3.74QAIAQDD427 pKa = 4.27LTWLGSISGIIKK439 pKa = 10.05ADD441 pKa = 3.49YY442 pKa = 9.7FQGGEE447 pKa = 4.01TAPQSMEE454 pKa = 3.67AAEE457 pKa = 4.56KK458 pKa = 10.47IEE460 pKa = 3.91PQVIDD465 pKa = 3.93EE466 pKa = 4.07VWPRR470 pKa = 11.84FTFTEE475 pKa = 4.12EE476 pKa = 3.75EE477 pKa = 3.93NRR479 pKa = 11.84ILQSTGEE486 pKa = 4.68DD487 pKa = 2.72INKK490 pKa = 9.52YY491 pKa = 8.28VEE493 pKa = 3.8EE494 pKa = 4.26MRR496 pKa = 11.84DD497 pKa = 3.28KK498 pKa = 11.1FITGDD503 pKa = 3.6EE504 pKa = 4.73DD505 pKa = 4.33LDD507 pKa = 3.67NWDD510 pKa = 4.82SYY512 pKa = 11.15IQTIEE517 pKa = 3.82QMGMEE522 pKa = 4.66EE523 pKa = 4.08LQEE526 pKa = 4.38VYY528 pKa = 9.93QAAYY532 pKa = 10.44DD533 pKa = 4.14RR534 pKa = 11.84YY535 pKa = 10.37QSNN538 pKa = 3.03
MM1 pKa = 7.18LKK3 pKa = 9.95KK4 pKa = 9.75WFFFIATLVFVSALYY19 pKa = 10.47ACSGGDD25 pKa = 4.17DD26 pKa = 3.8GDD28 pKa = 4.2SKK30 pKa = 11.1EE31 pKa = 4.22GEE33 pKa = 4.08AAGGDD38 pKa = 3.74NVNATGFPIVEE49 pKa = 4.49EE50 pKa = 4.05EE51 pKa = 4.42LEE53 pKa = 4.1LTIFANKK60 pKa = 8.56PAQNEE65 pKa = 4.15GNDD68 pKa = 3.57WNDD71 pKa = 2.75ILIWNTYY78 pKa = 10.16RR79 pKa = 11.84DD80 pKa = 3.75LTNINVNWNSVSPDD94 pKa = 3.09ALEE97 pKa = 4.15EE98 pKa = 3.98NRR100 pKa = 11.84NLALGSGDD108 pKa = 4.09LPDD111 pKa = 4.49AFFLAEE117 pKa = 4.79LSNTDD122 pKa = 3.68LLRR125 pKa = 11.84YY126 pKa = 9.34GSQGAFLPLNDD137 pKa = 5.84LIDD140 pKa = 4.31EE141 pKa = 4.35YY142 pKa = 11.74APNLKK147 pKa = 10.42ALMEE151 pKa = 4.59NDD153 pKa = 3.42PTIEE157 pKa = 4.04KK158 pKa = 10.55AITFPDD164 pKa = 3.68GNIYY168 pKa = 10.87SMPSLIEE175 pKa = 4.77DD176 pKa = 4.05DD177 pKa = 4.36FLSLRR182 pKa = 11.84LSARR186 pKa = 11.84PWVNEE191 pKa = 3.52DD192 pKa = 3.02WLNEE196 pKa = 3.75LDD198 pKa = 3.64MDD200 pKa = 4.28IPEE203 pKa = 4.45TTGDD207 pKa = 3.31FYY209 pKa = 11.51EE210 pKa = 4.13YY211 pKa = 10.74LKK213 pKa = 10.83AVKK216 pKa = 10.14EE217 pKa = 3.99LDD219 pKa = 3.43PVGNGDD225 pKa = 4.24TIPYY229 pKa = 9.4GGTDD233 pKa = 2.94IAEE236 pKa = 4.22LVQWLSGSFGVMNHH250 pKa = 6.26GASNTNLDD258 pKa = 4.69LDD260 pKa = 4.75PNDD263 pKa = 3.95DD264 pKa = 3.8SKK266 pKa = 11.74VRR268 pKa = 11.84YY269 pKa = 8.93YY270 pKa = 10.46ATADD274 pKa = 3.45EE275 pKa = 4.16YY276 pKa = 11.48RR277 pKa = 11.84EE278 pKa = 3.92MLEE281 pKa = 4.31YY282 pKa = 10.42IHH284 pKa = 6.45MLYY287 pKa = 10.67EE288 pKa = 4.35EE289 pKa = 4.44GLIDD293 pKa = 4.2QSIFTIEE300 pKa = 3.65WGQFLANASDD310 pKa = 3.62NLYY313 pKa = 11.17GSMIFYY319 pKa = 10.71DD320 pKa = 4.5PIEE323 pKa = 4.34LFGEE327 pKa = 4.68EE328 pKa = 4.82IGSQYY333 pKa = 11.39NSLAALEE340 pKa = 5.1GPDD343 pKa = 3.69GHH345 pKa = 6.63QSYY348 pKa = 11.15NKK350 pKa = 9.86LSSSVWTPDD359 pKa = 2.93NLVITSEE366 pKa = 4.25NPNPAATVRR375 pKa = 11.84WMDD378 pKa = 3.36HH379 pKa = 6.28FYY381 pKa = 11.23GDD383 pKa = 3.91EE384 pKa = 4.15GAEE387 pKa = 3.99LYY389 pKa = 11.42YY390 pKa = 10.25MGVEE394 pKa = 4.09GEE396 pKa = 4.28TFEE399 pKa = 5.76IEE401 pKa = 4.17DD402 pKa = 4.0GEE404 pKa = 4.55AVYY407 pKa = 10.79SDD409 pKa = 5.26HH410 pKa = 7.0ILNPEE415 pKa = 3.49GDD417 pKa = 3.72MTFEE421 pKa = 3.74QAIAQDD427 pKa = 4.27LTWLGSISGIIKK439 pKa = 10.05ADD441 pKa = 3.49YY442 pKa = 9.7FQGGEE447 pKa = 4.01TAPQSMEE454 pKa = 3.67AAEE457 pKa = 4.56KK458 pKa = 10.47IEE460 pKa = 3.91PQVIDD465 pKa = 3.93EE466 pKa = 4.07VWPRR470 pKa = 11.84FTFTEE475 pKa = 4.12EE476 pKa = 3.75EE477 pKa = 3.93NRR479 pKa = 11.84ILQSTGEE486 pKa = 4.68DD487 pKa = 2.72INKK490 pKa = 9.52YY491 pKa = 8.28VEE493 pKa = 3.8EE494 pKa = 4.26MRR496 pKa = 11.84DD497 pKa = 3.28KK498 pKa = 11.1FITGDD503 pKa = 3.6EE504 pKa = 4.73DD505 pKa = 4.33LDD507 pKa = 3.67NWDD510 pKa = 4.82SYY512 pKa = 11.15IQTIEE517 pKa = 3.82QMGMEE522 pKa = 4.66EE523 pKa = 4.08LQEE526 pKa = 4.38VYY528 pKa = 9.93QAAYY532 pKa = 10.44DD533 pKa = 4.14RR534 pKa = 11.84YY535 pKa = 10.37QSNN538 pKa = 3.03
Molecular weight: 60.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A1M526|A0A0A1M526_9BACI HTH-type transcriptional regulator CynR OS=Oceanobacillus oncorhynchi OX=545501 GN=cynR_2 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 9.9NGRR28 pKa = 11.84HH29 pKa = 4.47ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.09GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.46VHH16 pKa = 5.5GFRR19 pKa = 11.84TRR21 pKa = 11.84MSTKK25 pKa = 9.9NGRR28 pKa = 11.84HH29 pKa = 4.47ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.09GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1268742 |
29 |
2686 |
289.3 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.418 ± 0.038 | 0.647 ± 0.011 |
5.326 ± 0.033 | 7.952 ± 0.053 |
4.547 ± 0.033 | 6.835 ± 0.039 |
2.069 ± 0.022 | 8.107 ± 0.038 |
6.373 ± 0.042 | 9.561 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.88 ± 0.019 | 4.344 ± 0.027 |
3.516 ± 0.019 | 3.95 ± 0.03 |
3.821 ± 0.027 | 5.917 ± 0.026 |
5.382 ± 0.023 | 6.786 ± 0.031 |
1.026 ± 0.016 | 3.542 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
