
Beihai narna-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
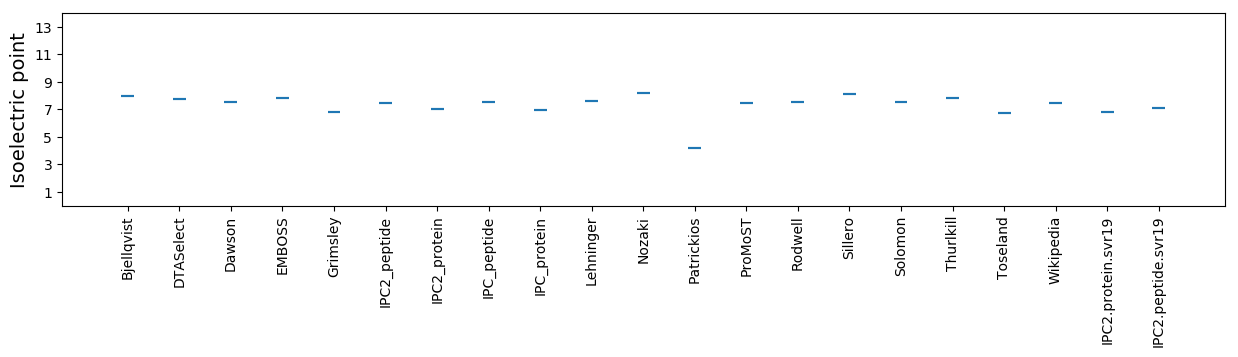
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KI50|A0A1L3KI50_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 4 OX=1922456 PE=4 SV=1
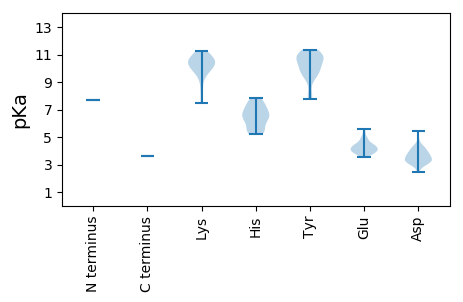
MM1 pKa = 7.66PVLSTLRR8 pKa = 11.84IACVGYY14 pKa = 10.7DD15 pKa = 3.33PTKK18 pKa = 10.64EE19 pKa = 4.0YY20 pKa = 11.14LEE22 pKa = 3.98QALAEE27 pKa = 4.24RR28 pKa = 11.84TARR31 pKa = 11.84MAGDD35 pKa = 4.2IPCASDD41 pKa = 3.19VDD43 pKa = 4.18EE44 pKa = 5.61SSDD47 pKa = 4.08GPGVCADD54 pKa = 4.34AYY56 pKa = 10.88LHH58 pKa = 7.4GIRR61 pKa = 11.84QMDD64 pKa = 3.57RR65 pKa = 11.84VEE67 pKa = 4.46EE68 pKa = 4.38VWSHH72 pKa = 6.58IYY74 pKa = 9.71RR75 pKa = 11.84CYY77 pKa = 10.69EE78 pKa = 3.56IDD80 pKa = 3.3PTPFSEE86 pKa = 4.25RR87 pKa = 11.84DD88 pKa = 2.98RR89 pKa = 11.84GLFLWLGAHH98 pKa = 6.65DD99 pKa = 3.96QLEE102 pKa = 4.84SYY104 pKa = 10.17LKK106 pKa = 10.26WRR108 pKa = 11.84TAWLLAQVWEE118 pKa = 4.14QRR120 pKa = 11.84EE121 pKa = 4.38LPSKK125 pKa = 10.29PGYY128 pKa = 10.12LSEE131 pKa = 4.58VPNCYY136 pKa = 9.79RR137 pKa = 11.84PPRR140 pKa = 11.84VGLTWGDD147 pKa = 3.49SAFWQRR153 pKa = 11.84AIHH156 pKa = 5.53QSGNIPLRR164 pKa = 11.84LQEE167 pKa = 4.26CAYY170 pKa = 10.84ALYY173 pKa = 10.05QGKK176 pKa = 10.2AGALPMRR183 pKa = 11.84PDD185 pKa = 3.3LVDD188 pKa = 3.75AKK190 pKa = 11.03VEE192 pKa = 4.03EE193 pKa = 4.45AMSRR197 pKa = 11.84LTTPCRR203 pKa = 11.84TRR205 pKa = 11.84SQVIGGRR212 pKa = 11.84KK213 pKa = 7.47VTLQDD218 pKa = 3.38LKK220 pKa = 10.73EE221 pKa = 4.11QVTRR225 pKa = 11.84TVKK228 pKa = 10.45EE229 pKa = 3.64IYY231 pKa = 9.44GPSMEE236 pKa = 4.92EE237 pKa = 3.79EE238 pKa = 4.41VPTPGSHH245 pKa = 7.27RR246 pKa = 11.84LGSTKK251 pKa = 10.48SSFQCPRR258 pKa = 11.84GGGGAHH264 pKa = 5.39QALVEE269 pKa = 4.1EE270 pKa = 4.78AGEE273 pKa = 4.23GRR275 pKa = 11.84AGVLGLPQLVGYY287 pKa = 7.73TRR289 pKa = 11.84EE290 pKa = 3.9RR291 pKa = 11.84DD292 pKa = 3.64PVPVYY297 pKa = 10.24STTDD301 pKa = 3.13HH302 pKa = 6.88LEE304 pKa = 3.69WSHH307 pKa = 5.51TVEE310 pKa = 3.82ASKK313 pKa = 11.0VLSFGEE319 pKa = 4.31TMACYY324 pKa = 9.32PVGLVEE330 pKa = 4.06PFKK333 pKa = 11.27VRR335 pKa = 11.84VITRR339 pKa = 11.84GAAHH343 pKa = 7.39HH344 pKa = 5.49YY345 pKa = 9.71HH346 pKa = 7.26LARR349 pKa = 11.84RR350 pKa = 11.84WQPSMWRR357 pKa = 11.84PLAAHH362 pKa = 6.54PTFQLVGRR370 pKa = 11.84PCGRR374 pKa = 11.84DD375 pKa = 3.17VMYY378 pKa = 10.99DD379 pKa = 3.29LVRR382 pKa = 11.84KK383 pKa = 9.99VDD385 pKa = 4.08FGDD388 pKa = 3.24GRR390 pKa = 11.84LWMSGDD396 pKa = 3.51YY397 pKa = 10.04QAATDD402 pKa = 3.96YY403 pKa = 10.98FDD405 pKa = 5.43PEE407 pKa = 4.39LSGHH411 pKa = 6.36CLMEE415 pKa = 4.41VCDD418 pKa = 4.11SLGVPWEE425 pKa = 3.86DD426 pKa = 3.38RR427 pKa = 11.84IVLRR431 pKa = 11.84RR432 pKa = 11.84ALIGHH437 pKa = 6.29VFHH440 pKa = 7.84DD441 pKa = 4.77PDD443 pKa = 3.3THH445 pKa = 6.9EE446 pKa = 4.38YY447 pKa = 10.5LGHH450 pKa = 5.19QARR453 pKa = 11.84GQLMGSPVSFPILCLLNAALTRR475 pKa = 11.84FAFEE479 pKa = 3.76LAGRR483 pKa = 11.84EE484 pKa = 3.85ASMLRR489 pKa = 11.84DD490 pKa = 3.34QPILVNGDD498 pKa = 3.88DD499 pKa = 4.15LLCRR503 pKa = 11.84TSDD506 pKa = 3.04KK507 pKa = 10.87EE508 pKa = 4.09YY509 pKa = 10.73YY510 pKa = 9.33IWSQITAMGGLIPSLGKK527 pKa = 10.18CFRR530 pKa = 11.84HH531 pKa = 6.41RR532 pKa = 11.84RR533 pKa = 11.84IATINSEE540 pKa = 3.72MWVCRR545 pKa = 11.84KK546 pKa = 8.49MRR548 pKa = 11.84TTLLGPLDD556 pKa = 4.31GPLSEE561 pKa = 4.56FTYY564 pKa = 8.21TTIEE568 pKa = 4.07RR569 pKa = 11.84RR570 pKa = 11.84HH571 pKa = 6.06LPLEE575 pKa = 4.11GLARR579 pKa = 11.84GSIKK583 pKa = 10.46GASGTTDD590 pKa = 2.81SQRR593 pKa = 11.84RR594 pKa = 11.84IEE596 pKa = 5.07KK597 pKa = 9.78MSLLDD602 pKa = 3.59SSNVRR607 pKa = 11.84SLVPMGDD614 pKa = 2.98CWRR617 pKa = 11.84EE618 pKa = 3.97YY619 pKa = 10.71VRR621 pKa = 11.84STPSYY626 pKa = 10.84ADD628 pKa = 3.22EE629 pKa = 4.44SSPGVRR635 pKa = 11.84AALQRR640 pKa = 11.84RR641 pKa = 11.84LWDD644 pKa = 3.27HH645 pKa = 6.27CWDD648 pKa = 3.7VNRR651 pKa = 11.84DD652 pKa = 3.24RR653 pKa = 11.84LKK655 pKa = 10.61RR656 pKa = 11.84ISRR659 pKa = 11.84FPISFCMPVRR669 pKa = 11.84YY670 pKa = 9.72GGLGFPLPPPSSRR683 pKa = 11.84SYY685 pKa = 10.97AQRR688 pKa = 11.84LPKK691 pKa = 10.17ANNWAMANLLANRR704 pKa = 11.84PNLRR708 pKa = 11.84RR709 pKa = 11.84ALLSQLKK716 pKa = 9.98PDD718 pKa = 3.69VPCYY722 pKa = 10.97GDD724 pKa = 2.96VAYY727 pKa = 9.58PLRR730 pKa = 11.84CVPNRR735 pKa = 11.84DD736 pKa = 3.37ANLDD740 pKa = 3.58EE741 pKa = 5.38EE742 pKa = 4.74YY743 pKa = 11.17DD744 pKa = 3.76SFRR747 pKa = 11.84RR748 pKa = 11.84SYY750 pKa = 11.03PDD752 pKa = 3.61LPPEE756 pKa = 4.03SFAPRR761 pKa = 11.84RR762 pKa = 11.84GLPSRR767 pKa = 11.84PSTDD771 pKa = 2.87PSFWGMCGSEE781 pKa = 3.77PTQYY785 pKa = 11.35YY786 pKa = 10.35EE787 pKa = 4.96DD788 pKa = 4.34DD789 pKa = 3.78SGFLRR794 pKa = 11.84IWNAANRR801 pKa = 11.84IAQSKK806 pKa = 9.53NSSGKK811 pKa = 9.4YY812 pKa = 7.88RR813 pKa = 11.84WAPLRR818 pKa = 11.84RR819 pKa = 11.84LSLLDD824 pKa = 3.87TIALSDD830 pKa = 3.78HH831 pKa = 6.7KK832 pKa = 10.73LTGLDD837 pKa = 4.42LPHH840 pKa = 7.71SDD842 pKa = 2.42IRR844 pKa = 11.84SPFRR848 pKa = 11.84DD849 pKa = 3.15TGVSRR854 pKa = 11.84HH855 pKa = 5.37EE856 pKa = 4.01VAALGDD862 pKa = 3.78VVLNEE867 pKa = 4.11LWW869 pKa = 3.59
MM1 pKa = 7.66PVLSTLRR8 pKa = 11.84IACVGYY14 pKa = 10.7DD15 pKa = 3.33PTKK18 pKa = 10.64EE19 pKa = 4.0YY20 pKa = 11.14LEE22 pKa = 3.98QALAEE27 pKa = 4.24RR28 pKa = 11.84TARR31 pKa = 11.84MAGDD35 pKa = 4.2IPCASDD41 pKa = 3.19VDD43 pKa = 4.18EE44 pKa = 5.61SSDD47 pKa = 4.08GPGVCADD54 pKa = 4.34AYY56 pKa = 10.88LHH58 pKa = 7.4GIRR61 pKa = 11.84QMDD64 pKa = 3.57RR65 pKa = 11.84VEE67 pKa = 4.46EE68 pKa = 4.38VWSHH72 pKa = 6.58IYY74 pKa = 9.71RR75 pKa = 11.84CYY77 pKa = 10.69EE78 pKa = 3.56IDD80 pKa = 3.3PTPFSEE86 pKa = 4.25RR87 pKa = 11.84DD88 pKa = 2.98RR89 pKa = 11.84GLFLWLGAHH98 pKa = 6.65DD99 pKa = 3.96QLEE102 pKa = 4.84SYY104 pKa = 10.17LKK106 pKa = 10.26WRR108 pKa = 11.84TAWLLAQVWEE118 pKa = 4.14QRR120 pKa = 11.84EE121 pKa = 4.38LPSKK125 pKa = 10.29PGYY128 pKa = 10.12LSEE131 pKa = 4.58VPNCYY136 pKa = 9.79RR137 pKa = 11.84PPRR140 pKa = 11.84VGLTWGDD147 pKa = 3.49SAFWQRR153 pKa = 11.84AIHH156 pKa = 5.53QSGNIPLRR164 pKa = 11.84LQEE167 pKa = 4.26CAYY170 pKa = 10.84ALYY173 pKa = 10.05QGKK176 pKa = 10.2AGALPMRR183 pKa = 11.84PDD185 pKa = 3.3LVDD188 pKa = 3.75AKK190 pKa = 11.03VEE192 pKa = 4.03EE193 pKa = 4.45AMSRR197 pKa = 11.84LTTPCRR203 pKa = 11.84TRR205 pKa = 11.84SQVIGGRR212 pKa = 11.84KK213 pKa = 7.47VTLQDD218 pKa = 3.38LKK220 pKa = 10.73EE221 pKa = 4.11QVTRR225 pKa = 11.84TVKK228 pKa = 10.45EE229 pKa = 3.64IYY231 pKa = 9.44GPSMEE236 pKa = 4.92EE237 pKa = 3.79EE238 pKa = 4.41VPTPGSHH245 pKa = 7.27RR246 pKa = 11.84LGSTKK251 pKa = 10.48SSFQCPRR258 pKa = 11.84GGGGAHH264 pKa = 5.39QALVEE269 pKa = 4.1EE270 pKa = 4.78AGEE273 pKa = 4.23GRR275 pKa = 11.84AGVLGLPQLVGYY287 pKa = 7.73TRR289 pKa = 11.84EE290 pKa = 3.9RR291 pKa = 11.84DD292 pKa = 3.64PVPVYY297 pKa = 10.24STTDD301 pKa = 3.13HH302 pKa = 6.88LEE304 pKa = 3.69WSHH307 pKa = 5.51TVEE310 pKa = 3.82ASKK313 pKa = 11.0VLSFGEE319 pKa = 4.31TMACYY324 pKa = 9.32PVGLVEE330 pKa = 4.06PFKK333 pKa = 11.27VRR335 pKa = 11.84VITRR339 pKa = 11.84GAAHH343 pKa = 7.39HH344 pKa = 5.49YY345 pKa = 9.71HH346 pKa = 7.26LARR349 pKa = 11.84RR350 pKa = 11.84WQPSMWRR357 pKa = 11.84PLAAHH362 pKa = 6.54PTFQLVGRR370 pKa = 11.84PCGRR374 pKa = 11.84DD375 pKa = 3.17VMYY378 pKa = 10.99DD379 pKa = 3.29LVRR382 pKa = 11.84KK383 pKa = 9.99VDD385 pKa = 4.08FGDD388 pKa = 3.24GRR390 pKa = 11.84LWMSGDD396 pKa = 3.51YY397 pKa = 10.04QAATDD402 pKa = 3.96YY403 pKa = 10.98FDD405 pKa = 5.43PEE407 pKa = 4.39LSGHH411 pKa = 6.36CLMEE415 pKa = 4.41VCDD418 pKa = 4.11SLGVPWEE425 pKa = 3.86DD426 pKa = 3.38RR427 pKa = 11.84IVLRR431 pKa = 11.84RR432 pKa = 11.84ALIGHH437 pKa = 6.29VFHH440 pKa = 7.84DD441 pKa = 4.77PDD443 pKa = 3.3THH445 pKa = 6.9EE446 pKa = 4.38YY447 pKa = 10.5LGHH450 pKa = 5.19QARR453 pKa = 11.84GQLMGSPVSFPILCLLNAALTRR475 pKa = 11.84FAFEE479 pKa = 3.76LAGRR483 pKa = 11.84EE484 pKa = 3.85ASMLRR489 pKa = 11.84DD490 pKa = 3.34QPILVNGDD498 pKa = 3.88DD499 pKa = 4.15LLCRR503 pKa = 11.84TSDD506 pKa = 3.04KK507 pKa = 10.87EE508 pKa = 4.09YY509 pKa = 10.73YY510 pKa = 9.33IWSQITAMGGLIPSLGKK527 pKa = 10.18CFRR530 pKa = 11.84HH531 pKa = 6.41RR532 pKa = 11.84RR533 pKa = 11.84IATINSEE540 pKa = 3.72MWVCRR545 pKa = 11.84KK546 pKa = 8.49MRR548 pKa = 11.84TTLLGPLDD556 pKa = 4.31GPLSEE561 pKa = 4.56FTYY564 pKa = 8.21TTIEE568 pKa = 4.07RR569 pKa = 11.84RR570 pKa = 11.84HH571 pKa = 6.06LPLEE575 pKa = 4.11GLARR579 pKa = 11.84GSIKK583 pKa = 10.46GASGTTDD590 pKa = 2.81SQRR593 pKa = 11.84RR594 pKa = 11.84IEE596 pKa = 5.07KK597 pKa = 9.78MSLLDD602 pKa = 3.59SSNVRR607 pKa = 11.84SLVPMGDD614 pKa = 2.98CWRR617 pKa = 11.84EE618 pKa = 3.97YY619 pKa = 10.71VRR621 pKa = 11.84STPSYY626 pKa = 10.84ADD628 pKa = 3.22EE629 pKa = 4.44SSPGVRR635 pKa = 11.84AALQRR640 pKa = 11.84RR641 pKa = 11.84LWDD644 pKa = 3.27HH645 pKa = 6.27CWDD648 pKa = 3.7VNRR651 pKa = 11.84DD652 pKa = 3.24RR653 pKa = 11.84LKK655 pKa = 10.61RR656 pKa = 11.84ISRR659 pKa = 11.84FPISFCMPVRR669 pKa = 11.84YY670 pKa = 9.72GGLGFPLPPPSSRR683 pKa = 11.84SYY685 pKa = 10.97AQRR688 pKa = 11.84LPKK691 pKa = 10.17ANNWAMANLLANRR704 pKa = 11.84PNLRR708 pKa = 11.84RR709 pKa = 11.84ALLSQLKK716 pKa = 9.98PDD718 pKa = 3.69VPCYY722 pKa = 10.97GDD724 pKa = 2.96VAYY727 pKa = 9.58PLRR730 pKa = 11.84CVPNRR735 pKa = 11.84DD736 pKa = 3.37ANLDD740 pKa = 3.58EE741 pKa = 5.38EE742 pKa = 4.74YY743 pKa = 11.17DD744 pKa = 3.76SFRR747 pKa = 11.84RR748 pKa = 11.84SYY750 pKa = 11.03PDD752 pKa = 3.61LPPEE756 pKa = 4.03SFAPRR761 pKa = 11.84RR762 pKa = 11.84GLPSRR767 pKa = 11.84PSTDD771 pKa = 2.87PSFWGMCGSEE781 pKa = 3.77PTQYY785 pKa = 11.35YY786 pKa = 10.35EE787 pKa = 4.96DD788 pKa = 4.34DD789 pKa = 3.78SGFLRR794 pKa = 11.84IWNAANRR801 pKa = 11.84IAQSKK806 pKa = 9.53NSSGKK811 pKa = 9.4YY812 pKa = 7.88RR813 pKa = 11.84WAPLRR818 pKa = 11.84RR819 pKa = 11.84LSLLDD824 pKa = 3.87TIALSDD830 pKa = 3.78HH831 pKa = 6.7KK832 pKa = 10.73LTGLDD837 pKa = 4.42LPHH840 pKa = 7.71SDD842 pKa = 2.42IRR844 pKa = 11.84SPFRR848 pKa = 11.84DD849 pKa = 3.15TGVSRR854 pKa = 11.84HH855 pKa = 5.37EE856 pKa = 4.01VAALGDD862 pKa = 3.78VVLNEE867 pKa = 4.11LWW869 pKa = 3.59
Molecular weight: 98.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KI50|A0A1L3KI50_9VIRU RNA-dependent RNA polymerase OS=Beihai narna-like virus 4 OX=1922456 PE=4 SV=1
MM1 pKa = 7.66PVLSTLRR8 pKa = 11.84IACVGYY14 pKa = 10.7DD15 pKa = 3.33PTKK18 pKa = 10.64EE19 pKa = 4.0YY20 pKa = 11.14LEE22 pKa = 3.98QALAEE27 pKa = 4.24RR28 pKa = 11.84TARR31 pKa = 11.84MAGDD35 pKa = 4.2IPCASDD41 pKa = 3.19VDD43 pKa = 4.18EE44 pKa = 5.61SSDD47 pKa = 4.08GPGVCADD54 pKa = 4.34AYY56 pKa = 10.88LHH58 pKa = 7.4GIRR61 pKa = 11.84QMDD64 pKa = 3.57RR65 pKa = 11.84VEE67 pKa = 4.46EE68 pKa = 4.38VWSHH72 pKa = 6.58IYY74 pKa = 9.71RR75 pKa = 11.84CYY77 pKa = 10.69EE78 pKa = 3.56IDD80 pKa = 3.3PTPFSEE86 pKa = 4.25RR87 pKa = 11.84DD88 pKa = 2.98RR89 pKa = 11.84GLFLWLGAHH98 pKa = 6.65DD99 pKa = 3.96QLEE102 pKa = 4.84SYY104 pKa = 10.17LKK106 pKa = 10.26WRR108 pKa = 11.84TAWLLAQVWEE118 pKa = 4.14QRR120 pKa = 11.84EE121 pKa = 4.38LPSKK125 pKa = 10.29PGYY128 pKa = 10.12LSEE131 pKa = 4.58VPNCYY136 pKa = 9.79RR137 pKa = 11.84PPRR140 pKa = 11.84VGLTWGDD147 pKa = 3.49SAFWQRR153 pKa = 11.84AIHH156 pKa = 5.53QSGNIPLRR164 pKa = 11.84LQEE167 pKa = 4.26CAYY170 pKa = 10.84ALYY173 pKa = 10.05QGKK176 pKa = 10.2AGALPMRR183 pKa = 11.84PDD185 pKa = 3.3LVDD188 pKa = 3.75AKK190 pKa = 11.03VEE192 pKa = 4.03EE193 pKa = 4.45AMSRR197 pKa = 11.84LTTPCRR203 pKa = 11.84TRR205 pKa = 11.84SQVIGGRR212 pKa = 11.84KK213 pKa = 7.47VTLQDD218 pKa = 3.38LKK220 pKa = 10.73EE221 pKa = 4.11QVTRR225 pKa = 11.84TVKK228 pKa = 10.45EE229 pKa = 3.64IYY231 pKa = 9.44GPSMEE236 pKa = 4.92EE237 pKa = 3.79EE238 pKa = 4.41VPTPGSHH245 pKa = 7.27RR246 pKa = 11.84LGSTKK251 pKa = 10.48SSFQCPRR258 pKa = 11.84GGGGAHH264 pKa = 5.39QALVEE269 pKa = 4.1EE270 pKa = 4.78AGEE273 pKa = 4.23GRR275 pKa = 11.84AGVLGLPQLVGYY287 pKa = 7.73TRR289 pKa = 11.84EE290 pKa = 3.9RR291 pKa = 11.84DD292 pKa = 3.64PVPVYY297 pKa = 10.24STTDD301 pKa = 3.13HH302 pKa = 6.88LEE304 pKa = 3.69WSHH307 pKa = 5.51TVEE310 pKa = 3.82ASKK313 pKa = 11.0VLSFGEE319 pKa = 4.31TMACYY324 pKa = 9.32PVGLVEE330 pKa = 4.06PFKK333 pKa = 11.27VRR335 pKa = 11.84VITRR339 pKa = 11.84GAAHH343 pKa = 7.39HH344 pKa = 5.49YY345 pKa = 9.71HH346 pKa = 7.26LARR349 pKa = 11.84RR350 pKa = 11.84WQPSMWRR357 pKa = 11.84PLAAHH362 pKa = 6.54PTFQLVGRR370 pKa = 11.84PCGRR374 pKa = 11.84DD375 pKa = 3.17VMYY378 pKa = 10.99DD379 pKa = 3.29LVRR382 pKa = 11.84KK383 pKa = 9.99VDD385 pKa = 4.08FGDD388 pKa = 3.24GRR390 pKa = 11.84LWMSGDD396 pKa = 3.51YY397 pKa = 10.04QAATDD402 pKa = 3.96YY403 pKa = 10.98FDD405 pKa = 5.43PEE407 pKa = 4.39LSGHH411 pKa = 6.36CLMEE415 pKa = 4.41VCDD418 pKa = 4.11SLGVPWEE425 pKa = 3.86DD426 pKa = 3.38RR427 pKa = 11.84IVLRR431 pKa = 11.84RR432 pKa = 11.84ALIGHH437 pKa = 6.29VFHH440 pKa = 7.84DD441 pKa = 4.77PDD443 pKa = 3.3THH445 pKa = 6.9EE446 pKa = 4.38YY447 pKa = 10.5LGHH450 pKa = 5.19QARR453 pKa = 11.84GQLMGSPVSFPILCLLNAALTRR475 pKa = 11.84FAFEE479 pKa = 3.76LAGRR483 pKa = 11.84EE484 pKa = 3.85ASMLRR489 pKa = 11.84DD490 pKa = 3.34QPILVNGDD498 pKa = 3.88DD499 pKa = 4.15LLCRR503 pKa = 11.84TSDD506 pKa = 3.04KK507 pKa = 10.87EE508 pKa = 4.09YY509 pKa = 10.73YY510 pKa = 9.33IWSQITAMGGLIPSLGKK527 pKa = 10.18CFRR530 pKa = 11.84HH531 pKa = 6.41RR532 pKa = 11.84RR533 pKa = 11.84IATINSEE540 pKa = 3.72MWVCRR545 pKa = 11.84KK546 pKa = 8.49MRR548 pKa = 11.84TTLLGPLDD556 pKa = 4.31GPLSEE561 pKa = 4.56FTYY564 pKa = 8.21TTIEE568 pKa = 4.07RR569 pKa = 11.84RR570 pKa = 11.84HH571 pKa = 6.06LPLEE575 pKa = 4.11GLARR579 pKa = 11.84GSIKK583 pKa = 10.46GASGTTDD590 pKa = 2.81SQRR593 pKa = 11.84RR594 pKa = 11.84IEE596 pKa = 5.07KK597 pKa = 9.78MSLLDD602 pKa = 3.59SSNVRR607 pKa = 11.84SLVPMGDD614 pKa = 2.98CWRR617 pKa = 11.84EE618 pKa = 3.97YY619 pKa = 10.71VRR621 pKa = 11.84STPSYY626 pKa = 10.84ADD628 pKa = 3.22EE629 pKa = 4.44SSPGVRR635 pKa = 11.84AALQRR640 pKa = 11.84RR641 pKa = 11.84LWDD644 pKa = 3.27HH645 pKa = 6.27CWDD648 pKa = 3.7VNRR651 pKa = 11.84DD652 pKa = 3.24RR653 pKa = 11.84LKK655 pKa = 10.61RR656 pKa = 11.84ISRR659 pKa = 11.84FPISFCMPVRR669 pKa = 11.84YY670 pKa = 9.72GGLGFPLPPPSSRR683 pKa = 11.84SYY685 pKa = 10.97AQRR688 pKa = 11.84LPKK691 pKa = 10.17ANNWAMANLLANRR704 pKa = 11.84PNLRR708 pKa = 11.84RR709 pKa = 11.84ALLSQLKK716 pKa = 9.98PDD718 pKa = 3.69VPCYY722 pKa = 10.97GDD724 pKa = 2.96VAYY727 pKa = 9.58PLRR730 pKa = 11.84CVPNRR735 pKa = 11.84DD736 pKa = 3.37ANLDD740 pKa = 3.58EE741 pKa = 5.38EE742 pKa = 4.74YY743 pKa = 11.17DD744 pKa = 3.76SFRR747 pKa = 11.84RR748 pKa = 11.84SYY750 pKa = 11.03PDD752 pKa = 3.61LPPEE756 pKa = 4.03SFAPRR761 pKa = 11.84RR762 pKa = 11.84GLPSRR767 pKa = 11.84PSTDD771 pKa = 2.87PSFWGMCGSEE781 pKa = 3.77PTQYY785 pKa = 11.35YY786 pKa = 10.35EE787 pKa = 4.96DD788 pKa = 4.34DD789 pKa = 3.78SGFLRR794 pKa = 11.84IWNAANRR801 pKa = 11.84IAQSKK806 pKa = 9.53NSSGKK811 pKa = 9.4YY812 pKa = 7.88RR813 pKa = 11.84WAPLRR818 pKa = 11.84RR819 pKa = 11.84LSLLDD824 pKa = 3.87TIALSDD830 pKa = 3.78HH831 pKa = 6.7KK832 pKa = 10.73LTGLDD837 pKa = 4.42LPHH840 pKa = 7.71SDD842 pKa = 2.42IRR844 pKa = 11.84SPFRR848 pKa = 11.84DD849 pKa = 3.15TGVSRR854 pKa = 11.84HH855 pKa = 5.37EE856 pKa = 4.01VAALGDD862 pKa = 3.78VVLNEE867 pKa = 4.11LWW869 pKa = 3.59
MM1 pKa = 7.66PVLSTLRR8 pKa = 11.84IACVGYY14 pKa = 10.7DD15 pKa = 3.33PTKK18 pKa = 10.64EE19 pKa = 4.0YY20 pKa = 11.14LEE22 pKa = 3.98QALAEE27 pKa = 4.24RR28 pKa = 11.84TARR31 pKa = 11.84MAGDD35 pKa = 4.2IPCASDD41 pKa = 3.19VDD43 pKa = 4.18EE44 pKa = 5.61SSDD47 pKa = 4.08GPGVCADD54 pKa = 4.34AYY56 pKa = 10.88LHH58 pKa = 7.4GIRR61 pKa = 11.84QMDD64 pKa = 3.57RR65 pKa = 11.84VEE67 pKa = 4.46EE68 pKa = 4.38VWSHH72 pKa = 6.58IYY74 pKa = 9.71RR75 pKa = 11.84CYY77 pKa = 10.69EE78 pKa = 3.56IDD80 pKa = 3.3PTPFSEE86 pKa = 4.25RR87 pKa = 11.84DD88 pKa = 2.98RR89 pKa = 11.84GLFLWLGAHH98 pKa = 6.65DD99 pKa = 3.96QLEE102 pKa = 4.84SYY104 pKa = 10.17LKK106 pKa = 10.26WRR108 pKa = 11.84TAWLLAQVWEE118 pKa = 4.14QRR120 pKa = 11.84EE121 pKa = 4.38LPSKK125 pKa = 10.29PGYY128 pKa = 10.12LSEE131 pKa = 4.58VPNCYY136 pKa = 9.79RR137 pKa = 11.84PPRR140 pKa = 11.84VGLTWGDD147 pKa = 3.49SAFWQRR153 pKa = 11.84AIHH156 pKa = 5.53QSGNIPLRR164 pKa = 11.84LQEE167 pKa = 4.26CAYY170 pKa = 10.84ALYY173 pKa = 10.05QGKK176 pKa = 10.2AGALPMRR183 pKa = 11.84PDD185 pKa = 3.3LVDD188 pKa = 3.75AKK190 pKa = 11.03VEE192 pKa = 4.03EE193 pKa = 4.45AMSRR197 pKa = 11.84LTTPCRR203 pKa = 11.84TRR205 pKa = 11.84SQVIGGRR212 pKa = 11.84KK213 pKa = 7.47VTLQDD218 pKa = 3.38LKK220 pKa = 10.73EE221 pKa = 4.11QVTRR225 pKa = 11.84TVKK228 pKa = 10.45EE229 pKa = 3.64IYY231 pKa = 9.44GPSMEE236 pKa = 4.92EE237 pKa = 3.79EE238 pKa = 4.41VPTPGSHH245 pKa = 7.27RR246 pKa = 11.84LGSTKK251 pKa = 10.48SSFQCPRR258 pKa = 11.84GGGGAHH264 pKa = 5.39QALVEE269 pKa = 4.1EE270 pKa = 4.78AGEE273 pKa = 4.23GRR275 pKa = 11.84AGVLGLPQLVGYY287 pKa = 7.73TRR289 pKa = 11.84EE290 pKa = 3.9RR291 pKa = 11.84DD292 pKa = 3.64PVPVYY297 pKa = 10.24STTDD301 pKa = 3.13HH302 pKa = 6.88LEE304 pKa = 3.69WSHH307 pKa = 5.51TVEE310 pKa = 3.82ASKK313 pKa = 11.0VLSFGEE319 pKa = 4.31TMACYY324 pKa = 9.32PVGLVEE330 pKa = 4.06PFKK333 pKa = 11.27VRR335 pKa = 11.84VITRR339 pKa = 11.84GAAHH343 pKa = 7.39HH344 pKa = 5.49YY345 pKa = 9.71HH346 pKa = 7.26LARR349 pKa = 11.84RR350 pKa = 11.84WQPSMWRR357 pKa = 11.84PLAAHH362 pKa = 6.54PTFQLVGRR370 pKa = 11.84PCGRR374 pKa = 11.84DD375 pKa = 3.17VMYY378 pKa = 10.99DD379 pKa = 3.29LVRR382 pKa = 11.84KK383 pKa = 9.99VDD385 pKa = 4.08FGDD388 pKa = 3.24GRR390 pKa = 11.84LWMSGDD396 pKa = 3.51YY397 pKa = 10.04QAATDD402 pKa = 3.96YY403 pKa = 10.98FDD405 pKa = 5.43PEE407 pKa = 4.39LSGHH411 pKa = 6.36CLMEE415 pKa = 4.41VCDD418 pKa = 4.11SLGVPWEE425 pKa = 3.86DD426 pKa = 3.38RR427 pKa = 11.84IVLRR431 pKa = 11.84RR432 pKa = 11.84ALIGHH437 pKa = 6.29VFHH440 pKa = 7.84DD441 pKa = 4.77PDD443 pKa = 3.3THH445 pKa = 6.9EE446 pKa = 4.38YY447 pKa = 10.5LGHH450 pKa = 5.19QARR453 pKa = 11.84GQLMGSPVSFPILCLLNAALTRR475 pKa = 11.84FAFEE479 pKa = 3.76LAGRR483 pKa = 11.84EE484 pKa = 3.85ASMLRR489 pKa = 11.84DD490 pKa = 3.34QPILVNGDD498 pKa = 3.88DD499 pKa = 4.15LLCRR503 pKa = 11.84TSDD506 pKa = 3.04KK507 pKa = 10.87EE508 pKa = 4.09YY509 pKa = 10.73YY510 pKa = 9.33IWSQITAMGGLIPSLGKK527 pKa = 10.18CFRR530 pKa = 11.84HH531 pKa = 6.41RR532 pKa = 11.84RR533 pKa = 11.84IATINSEE540 pKa = 3.72MWVCRR545 pKa = 11.84KK546 pKa = 8.49MRR548 pKa = 11.84TTLLGPLDD556 pKa = 4.31GPLSEE561 pKa = 4.56FTYY564 pKa = 8.21TTIEE568 pKa = 4.07RR569 pKa = 11.84RR570 pKa = 11.84HH571 pKa = 6.06LPLEE575 pKa = 4.11GLARR579 pKa = 11.84GSIKK583 pKa = 10.46GASGTTDD590 pKa = 2.81SQRR593 pKa = 11.84RR594 pKa = 11.84IEE596 pKa = 5.07KK597 pKa = 9.78MSLLDD602 pKa = 3.59SSNVRR607 pKa = 11.84SLVPMGDD614 pKa = 2.98CWRR617 pKa = 11.84EE618 pKa = 3.97YY619 pKa = 10.71VRR621 pKa = 11.84STPSYY626 pKa = 10.84ADD628 pKa = 3.22EE629 pKa = 4.44SSPGVRR635 pKa = 11.84AALQRR640 pKa = 11.84RR641 pKa = 11.84LWDD644 pKa = 3.27HH645 pKa = 6.27CWDD648 pKa = 3.7VNRR651 pKa = 11.84DD652 pKa = 3.24RR653 pKa = 11.84LKK655 pKa = 10.61RR656 pKa = 11.84ISRR659 pKa = 11.84FPISFCMPVRR669 pKa = 11.84YY670 pKa = 9.72GGLGFPLPPPSSRR683 pKa = 11.84SYY685 pKa = 10.97AQRR688 pKa = 11.84LPKK691 pKa = 10.17ANNWAMANLLANRR704 pKa = 11.84PNLRR708 pKa = 11.84RR709 pKa = 11.84ALLSQLKK716 pKa = 9.98PDD718 pKa = 3.69VPCYY722 pKa = 10.97GDD724 pKa = 2.96VAYY727 pKa = 9.58PLRR730 pKa = 11.84CVPNRR735 pKa = 11.84DD736 pKa = 3.37ANLDD740 pKa = 3.58EE741 pKa = 5.38EE742 pKa = 4.74YY743 pKa = 11.17DD744 pKa = 3.76SFRR747 pKa = 11.84RR748 pKa = 11.84SYY750 pKa = 11.03PDD752 pKa = 3.61LPPEE756 pKa = 4.03SFAPRR761 pKa = 11.84RR762 pKa = 11.84GLPSRR767 pKa = 11.84PSTDD771 pKa = 2.87PSFWGMCGSEE781 pKa = 3.77PTQYY785 pKa = 11.35YY786 pKa = 10.35EE787 pKa = 4.96DD788 pKa = 4.34DD789 pKa = 3.78SGFLRR794 pKa = 11.84IWNAANRR801 pKa = 11.84IAQSKK806 pKa = 9.53NSSGKK811 pKa = 9.4YY812 pKa = 7.88RR813 pKa = 11.84WAPLRR818 pKa = 11.84RR819 pKa = 11.84LSLLDD824 pKa = 3.87TIALSDD830 pKa = 3.78HH831 pKa = 6.7KK832 pKa = 10.73LTGLDD837 pKa = 4.42LPHH840 pKa = 7.71SDD842 pKa = 2.42IRR844 pKa = 11.84SPFRR848 pKa = 11.84DD849 pKa = 3.15TGVSRR854 pKa = 11.84HH855 pKa = 5.37EE856 pKa = 4.01VAALGDD862 pKa = 3.78VVLNEE867 pKa = 4.11LWW869 pKa = 3.59
Molecular weight: 98.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
869 |
869 |
869 |
869.0 |
98.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.135 ± 0.0 | 2.532 ± 0.0 |
6.214 ± 0.0 | 5.639 ± 0.0 |
2.647 ± 0.0 | 7.595 ± 0.0 |
2.647 ± 0.0 | 3.222 ± 0.0 |
2.647 ± 0.0 | 10.587 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.417 ± 0.0 | 2.071 ± 0.0 |
7.365 ± 0.0 | 3.222 ± 0.0 |
9.551 ± 0.0 | 7.71 ± 0.0 |
4.603 ± 0.0 | 5.869 ± 0.0 |
2.532 ± 0.0 | 3.797 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
