
Phomopsis longicolla circular virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
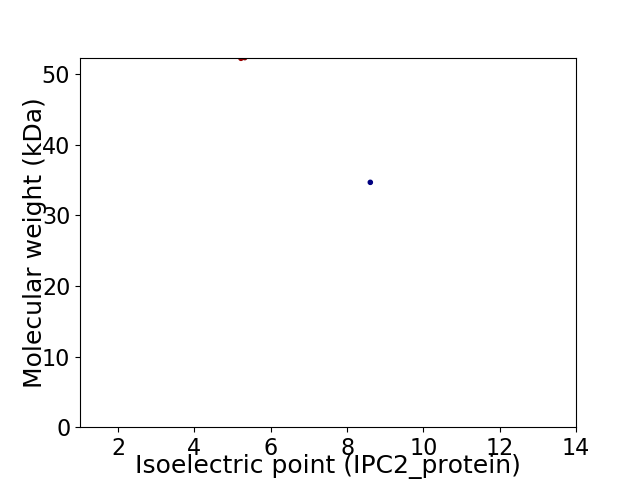
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S5QNP2|A0A1S5QNP2_9VIRU Putative coat protein OS=Phomopsis longicolla circular virus 1 OX=1808960 PE=4 SV=1
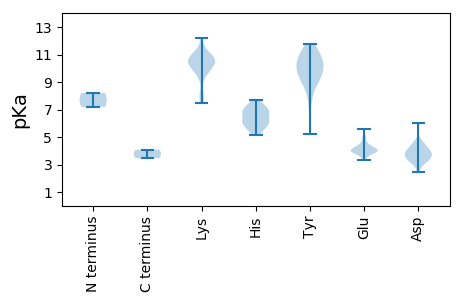
MM1 pKa = 8.2DD2 pKa = 4.6LTQLFLTSSPPPSHH16 pKa = 6.29QPNPLTLNIRR26 pKa = 11.84DD27 pKa = 4.1LSQDD31 pKa = 4.45DD32 pKa = 4.47FPQDD36 pKa = 3.69DD37 pKa = 3.91LSSFVVPDD45 pKa = 3.46SQFPGPEE52 pKa = 4.0ALPLSSPQFMTPLLHH67 pKa = 6.23TEE69 pKa = 4.3SLFVSQPDD77 pKa = 3.69EE78 pKa = 4.58DD79 pKa = 4.76DD80 pKa = 3.76QPEE83 pKa = 4.18DD84 pKa = 3.3QQDD87 pKa = 3.46AVGSDD92 pKa = 3.95DD93 pKa = 4.12EE94 pKa = 5.57LGLVDD99 pKa = 5.66PLDD102 pKa = 3.65EE103 pKa = 4.33TAGKK107 pKa = 9.76FRR109 pKa = 11.84LAAKK113 pKa = 10.05SVFLTYY119 pKa = 9.55PKK121 pKa = 10.34CHH123 pKa = 7.1LDD125 pKa = 3.17KK126 pKa = 11.06EE127 pKa = 4.82VFVKK131 pKa = 10.81AIEE134 pKa = 4.15KK135 pKa = 10.49FKK137 pKa = 10.86FDD139 pKa = 2.66QFYY142 pKa = 10.99AVRR145 pKa = 11.84EE146 pKa = 4.05DD147 pKa = 4.1HH148 pKa = 7.58KK149 pKa = 11.24DD150 pKa = 3.43GTPHH154 pKa = 5.17YY155 pKa = 9.51HH156 pKa = 7.2VIGEE160 pKa = 4.05WSVKK164 pKa = 10.54KK165 pKa = 10.4NIKK168 pKa = 9.77NPRR171 pKa = 11.84HH172 pKa = 5.88FDD174 pKa = 3.02IQKK177 pKa = 7.5YY178 pKa = 9.06HH179 pKa = 6.61PNIGRR184 pKa = 11.84TRR186 pKa = 11.84NRR188 pKa = 11.84LAAWRR193 pKa = 11.84YY194 pKa = 5.22VHH196 pKa = 5.81KK197 pKa = 9.66TKK199 pKa = 10.48GRR201 pKa = 11.84STHH204 pKa = 6.17VGGDD208 pKa = 3.22MPEE211 pKa = 3.8PVGIRR216 pKa = 11.84KK217 pKa = 7.79EE218 pKa = 4.13AKK220 pKa = 9.58DD221 pKa = 4.39DD222 pKa = 3.68FWKK225 pKa = 10.76EE226 pKa = 3.35AVTIKK231 pKa = 10.7DD232 pKa = 3.19RR233 pKa = 11.84NTFISRR239 pKa = 11.84FRR241 pKa = 11.84QEE243 pKa = 3.84APRR246 pKa = 11.84DD247 pKa = 4.0LIVHH251 pKa = 5.88YY252 pKa = 10.99SNVKK256 pKa = 10.3AYY258 pKa = 10.38ADD260 pKa = 3.92DD261 pKa = 3.92EE262 pKa = 5.08FKK264 pKa = 11.35VEE266 pKa = 3.91APEE269 pKa = 4.05YY270 pKa = 7.35CTPEE274 pKa = 3.62MSGYY278 pKa = 9.54WDD280 pKa = 3.84LPEE283 pKa = 4.38EE284 pKa = 3.98LKK286 pKa = 10.92EE287 pKa = 4.03WVEE290 pKa = 4.1EE291 pKa = 4.06NLTHH295 pKa = 7.66KK296 pKa = 10.29SARR299 pKa = 11.84PKK301 pKa = 10.48SLCLYY306 pKa = 10.94GEE308 pKa = 3.98TRR310 pKa = 11.84LYY312 pKa = 8.41KK313 pKa = 9.74TIWARR318 pKa = 11.84SLGPHH323 pKa = 6.62SYY325 pKa = 10.24MSGCWNARR333 pKa = 11.84LLDD336 pKa = 4.15DD337 pKa = 4.63DD338 pKa = 4.07KK339 pKa = 11.58QYY341 pKa = 11.79VVIDD345 pKa = 4.62DD346 pKa = 4.33VPLDD350 pKa = 4.1DD351 pKa = 5.99KK352 pKa = 12.17DD353 pKa = 3.64MFKK356 pKa = 10.63HH357 pKa = 5.86FKK359 pKa = 10.54QMLGCQNNFSVTDD372 pKa = 3.71KK373 pKa = 10.89YY374 pKa = 11.41VKK376 pKa = 10.13KK377 pKa = 10.59LHH379 pKa = 5.41FAKK382 pKa = 10.12WGLPCIYY389 pKa = 10.0LANQDD394 pKa = 3.36PRR396 pKa = 11.84EE397 pKa = 3.94YY398 pKa = 9.96STIDD402 pKa = 3.06GTHH405 pKa = 5.89RR406 pKa = 11.84RR407 pKa = 11.84WLNGNCVFVEE417 pKa = 4.17VEE419 pKa = 4.07KK420 pKa = 10.7PLFVASSGPGTPDD433 pKa = 2.49SRR435 pKa = 11.84QEE437 pKa = 3.9HH438 pKa = 6.8LSQFLQEE445 pKa = 4.4AEE447 pKa = 3.93QDD449 pKa = 3.44EE450 pKa = 5.0SFF452 pKa = 4.09
MM1 pKa = 8.2DD2 pKa = 4.6LTQLFLTSSPPPSHH16 pKa = 6.29QPNPLTLNIRR26 pKa = 11.84DD27 pKa = 4.1LSQDD31 pKa = 4.45DD32 pKa = 4.47FPQDD36 pKa = 3.69DD37 pKa = 3.91LSSFVVPDD45 pKa = 3.46SQFPGPEE52 pKa = 4.0ALPLSSPQFMTPLLHH67 pKa = 6.23TEE69 pKa = 4.3SLFVSQPDD77 pKa = 3.69EE78 pKa = 4.58DD79 pKa = 4.76DD80 pKa = 3.76QPEE83 pKa = 4.18DD84 pKa = 3.3QQDD87 pKa = 3.46AVGSDD92 pKa = 3.95DD93 pKa = 4.12EE94 pKa = 5.57LGLVDD99 pKa = 5.66PLDD102 pKa = 3.65EE103 pKa = 4.33TAGKK107 pKa = 9.76FRR109 pKa = 11.84LAAKK113 pKa = 10.05SVFLTYY119 pKa = 9.55PKK121 pKa = 10.34CHH123 pKa = 7.1LDD125 pKa = 3.17KK126 pKa = 11.06EE127 pKa = 4.82VFVKK131 pKa = 10.81AIEE134 pKa = 4.15KK135 pKa = 10.49FKK137 pKa = 10.86FDD139 pKa = 2.66QFYY142 pKa = 10.99AVRR145 pKa = 11.84EE146 pKa = 4.05DD147 pKa = 4.1HH148 pKa = 7.58KK149 pKa = 11.24DD150 pKa = 3.43GTPHH154 pKa = 5.17YY155 pKa = 9.51HH156 pKa = 7.2VIGEE160 pKa = 4.05WSVKK164 pKa = 10.54KK165 pKa = 10.4NIKK168 pKa = 9.77NPRR171 pKa = 11.84HH172 pKa = 5.88FDD174 pKa = 3.02IQKK177 pKa = 7.5YY178 pKa = 9.06HH179 pKa = 6.61PNIGRR184 pKa = 11.84TRR186 pKa = 11.84NRR188 pKa = 11.84LAAWRR193 pKa = 11.84YY194 pKa = 5.22VHH196 pKa = 5.81KK197 pKa = 9.66TKK199 pKa = 10.48GRR201 pKa = 11.84STHH204 pKa = 6.17VGGDD208 pKa = 3.22MPEE211 pKa = 3.8PVGIRR216 pKa = 11.84KK217 pKa = 7.79EE218 pKa = 4.13AKK220 pKa = 9.58DD221 pKa = 4.39DD222 pKa = 3.68FWKK225 pKa = 10.76EE226 pKa = 3.35AVTIKK231 pKa = 10.7DD232 pKa = 3.19RR233 pKa = 11.84NTFISRR239 pKa = 11.84FRR241 pKa = 11.84QEE243 pKa = 3.84APRR246 pKa = 11.84DD247 pKa = 4.0LIVHH251 pKa = 5.88YY252 pKa = 10.99SNVKK256 pKa = 10.3AYY258 pKa = 10.38ADD260 pKa = 3.92DD261 pKa = 3.92EE262 pKa = 5.08FKK264 pKa = 11.35VEE266 pKa = 3.91APEE269 pKa = 4.05YY270 pKa = 7.35CTPEE274 pKa = 3.62MSGYY278 pKa = 9.54WDD280 pKa = 3.84LPEE283 pKa = 4.38EE284 pKa = 3.98LKK286 pKa = 10.92EE287 pKa = 4.03WVEE290 pKa = 4.1EE291 pKa = 4.06NLTHH295 pKa = 7.66KK296 pKa = 10.29SARR299 pKa = 11.84PKK301 pKa = 10.48SLCLYY306 pKa = 10.94GEE308 pKa = 3.98TRR310 pKa = 11.84LYY312 pKa = 8.41KK313 pKa = 9.74TIWARR318 pKa = 11.84SLGPHH323 pKa = 6.62SYY325 pKa = 10.24MSGCWNARR333 pKa = 11.84LLDD336 pKa = 4.15DD337 pKa = 4.63DD338 pKa = 4.07KK339 pKa = 11.58QYY341 pKa = 11.79VVIDD345 pKa = 4.62DD346 pKa = 4.33VPLDD350 pKa = 4.1DD351 pKa = 5.99KK352 pKa = 12.17DD353 pKa = 3.64MFKK356 pKa = 10.63HH357 pKa = 5.86FKK359 pKa = 10.54QMLGCQNNFSVTDD372 pKa = 3.71KK373 pKa = 10.89YY374 pKa = 11.41VKK376 pKa = 10.13KK377 pKa = 10.59LHH379 pKa = 5.41FAKK382 pKa = 10.12WGLPCIYY389 pKa = 10.0LANQDD394 pKa = 3.36PRR396 pKa = 11.84EE397 pKa = 3.94YY398 pKa = 9.96STIDD402 pKa = 3.06GTHH405 pKa = 5.89RR406 pKa = 11.84RR407 pKa = 11.84WLNGNCVFVEE417 pKa = 4.17VEE419 pKa = 4.07KK420 pKa = 10.7PLFVASSGPGTPDD433 pKa = 2.49SRR435 pKa = 11.84QEE437 pKa = 3.9HH438 pKa = 6.8LSQFLQEE445 pKa = 4.4AEE447 pKa = 3.93QDD449 pKa = 3.44EE450 pKa = 5.0SFF452 pKa = 4.09
Molecular weight: 52.22 kDa
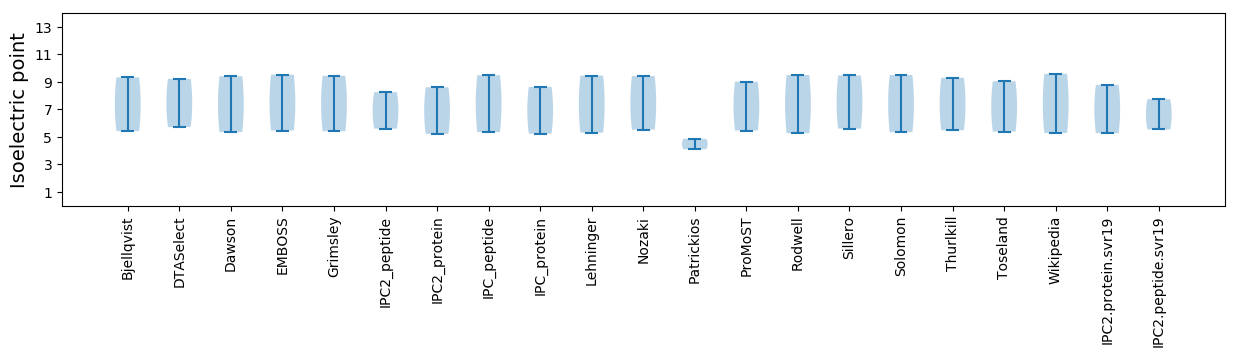
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S5QNP2|A0A1S5QNP2_9VIRU Putative coat protein OS=Phomopsis longicolla circular virus 1 OX=1808960 PE=4 SV=1
MM1 pKa = 7.19VFGRR5 pKa = 11.84RR6 pKa = 11.84GGYY9 pKa = 8.94RR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 6.63FYY14 pKa = 10.81NSRR17 pKa = 11.84PYY19 pKa = 10.89YY20 pKa = 10.29RR21 pKa = 11.84FASAVNSVGQEE32 pKa = 3.99KK33 pKa = 10.62KK34 pKa = 10.74NKK36 pKa = 8.04TVSLAGPGSASAHH49 pKa = 6.08CEE51 pKa = 3.77MHH53 pKa = 7.14AGDD56 pKa = 4.7PDD58 pKa = 3.9SGFVLPATEE67 pKa = 4.26VCRR70 pKa = 11.84SGDD73 pKa = 3.22RR74 pKa = 11.84SSGRR78 pKa = 11.84EE79 pKa = 3.58VHH81 pKa = 6.77LRR83 pKa = 11.84GAKK86 pKa = 9.95LSFEE90 pKa = 4.14ICVSRR95 pKa = 11.84TCSVRR100 pKa = 11.84VLSVVLDD107 pKa = 3.65TDD109 pKa = 4.34RR110 pKa = 11.84NWFNVTGDD118 pKa = 3.62SVGLKK123 pKa = 8.36NTQNGGFGPLSGTGHH138 pKa = 7.01GIVHH142 pKa = 6.86FLDD145 pKa = 3.62VNNGVFSSEE154 pKa = 4.47DD155 pKa = 3.14LRR157 pKa = 11.84PWFFRR162 pKa = 11.84SVSKK166 pKa = 9.7RR167 pKa = 11.84TKK169 pKa = 10.63GRR171 pKa = 11.84ILLDD175 pKa = 3.35QVQHH179 pKa = 5.23FTVGDD184 pKa = 3.91RR185 pKa = 11.84FQFRR189 pKa = 11.84PVSAWLDD196 pKa = 3.28LHH198 pKa = 7.04GKK200 pKa = 7.7VTYY203 pKa = 10.08QQTSDD208 pKa = 2.86IVDD211 pKa = 3.37RR212 pKa = 11.84DD213 pKa = 4.31GVCTRR218 pKa = 11.84GIDD221 pKa = 3.43EE222 pKa = 4.72KK223 pKa = 10.77PYY225 pKa = 9.39NVYY228 pKa = 10.6FIVLVHH234 pKa = 6.09VPSFSTEE241 pKa = 3.7QVVRR245 pKa = 11.84QSIGMPDD252 pKa = 3.9VGRR255 pKa = 11.84MDD257 pKa = 3.56VDD259 pKa = 3.78DD260 pKa = 4.34PRR262 pKa = 11.84VPKK265 pKa = 10.33RR266 pKa = 11.84EE267 pKa = 3.79RR268 pKa = 11.84GIPGYY273 pKa = 10.07GSRR276 pKa = 11.84PSTAEE281 pKa = 3.88SEE283 pKa = 4.28ASTIILDD290 pKa = 3.88RR291 pKa = 11.84KK292 pKa = 9.64GVAVPAIEE300 pKa = 3.94MRR302 pKa = 11.84NISSALYY309 pKa = 9.03WRR311 pKa = 11.84EE312 pKa = 3.46
MM1 pKa = 7.19VFGRR5 pKa = 11.84RR6 pKa = 11.84GGYY9 pKa = 8.94RR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 6.63FYY14 pKa = 10.81NSRR17 pKa = 11.84PYY19 pKa = 10.89YY20 pKa = 10.29RR21 pKa = 11.84FASAVNSVGQEE32 pKa = 3.99KK33 pKa = 10.62KK34 pKa = 10.74NKK36 pKa = 8.04TVSLAGPGSASAHH49 pKa = 6.08CEE51 pKa = 3.77MHH53 pKa = 7.14AGDD56 pKa = 4.7PDD58 pKa = 3.9SGFVLPATEE67 pKa = 4.26VCRR70 pKa = 11.84SGDD73 pKa = 3.22RR74 pKa = 11.84SSGRR78 pKa = 11.84EE79 pKa = 3.58VHH81 pKa = 6.77LRR83 pKa = 11.84GAKK86 pKa = 9.95LSFEE90 pKa = 4.14ICVSRR95 pKa = 11.84TCSVRR100 pKa = 11.84VLSVVLDD107 pKa = 3.65TDD109 pKa = 4.34RR110 pKa = 11.84NWFNVTGDD118 pKa = 3.62SVGLKK123 pKa = 8.36NTQNGGFGPLSGTGHH138 pKa = 7.01GIVHH142 pKa = 6.86FLDD145 pKa = 3.62VNNGVFSSEE154 pKa = 4.47DD155 pKa = 3.14LRR157 pKa = 11.84PWFFRR162 pKa = 11.84SVSKK166 pKa = 9.7RR167 pKa = 11.84TKK169 pKa = 10.63GRR171 pKa = 11.84ILLDD175 pKa = 3.35QVQHH179 pKa = 5.23FTVGDD184 pKa = 3.91RR185 pKa = 11.84FQFRR189 pKa = 11.84PVSAWLDD196 pKa = 3.28LHH198 pKa = 7.04GKK200 pKa = 7.7VTYY203 pKa = 10.08QQTSDD208 pKa = 2.86IVDD211 pKa = 3.37RR212 pKa = 11.84DD213 pKa = 4.31GVCTRR218 pKa = 11.84GIDD221 pKa = 3.43EE222 pKa = 4.72KK223 pKa = 10.77PYY225 pKa = 9.39NVYY228 pKa = 10.6FIVLVHH234 pKa = 6.09VPSFSTEE241 pKa = 3.7QVVRR245 pKa = 11.84QSIGMPDD252 pKa = 3.9VGRR255 pKa = 11.84MDD257 pKa = 3.56VDD259 pKa = 3.78DD260 pKa = 4.34PRR262 pKa = 11.84VPKK265 pKa = 10.33RR266 pKa = 11.84EE267 pKa = 3.79RR268 pKa = 11.84GIPGYY273 pKa = 10.07GSRR276 pKa = 11.84PSTAEE281 pKa = 3.88SEE283 pKa = 4.28ASTIILDD290 pKa = 3.88RR291 pKa = 11.84KK292 pKa = 9.64GVAVPAIEE300 pKa = 3.94MRR302 pKa = 11.84NISSALYY309 pKa = 9.03WRR311 pKa = 11.84EE312 pKa = 3.46
Molecular weight: 34.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
764 |
312 |
452 |
382.0 |
43.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.712 ± 0.132 | 1.571 ± 0.019 |
8.115 ± 0.999 | 5.759 ± 0.933 |
5.366 ± 0.14 | 6.806 ± 1.833 |
3.403 ± 0.304 | 3.403 ± 0.26 |
5.759 ± 1.308 | 7.199 ± 1.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.571 ± 0.019 | 3.403 ± 0.072 |
6.283 ± 0.864 | 3.927 ± 0.61 |
6.806 ± 1.645 | 7.984 ± 1.143 |
4.712 ± 0.056 | 8.246 ± 1.741 |
1.702 ± 0.246 | 3.272 ± 0.227 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
