
Mariniflexile fucanivorans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mariniflexile
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
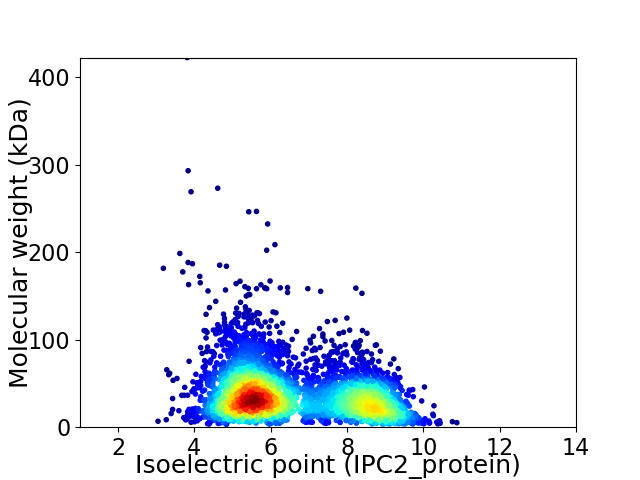
Virtual 2D-PAGE plot for 3991 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R1R972|A0A4R1R972_9FLAO Thiol peroxidase OS=Mariniflexile fucanivorans OX=264023 GN=tpx PE=3 SV=1
MM1 pKa = 7.54NLRR4 pKa = 11.84KK5 pKa = 9.85LSLYY9 pKa = 9.92LLCLTIGIVSCKK21 pKa = 10.04KK22 pKa = 10.52DD23 pKa = 3.71DD24 pKa = 4.61DD25 pKa = 4.03PTTVLVEE32 pKa = 3.85IEE34 pKa = 4.47DD35 pKa = 4.57RR36 pKa = 11.84DD37 pKa = 4.02VQQLKK42 pKa = 11.21DD43 pKa = 3.43MDD45 pKa = 4.64SLNKK49 pKa = 10.44YY50 pKa = 10.61LDD52 pKa = 3.23THH54 pKa = 6.69YY55 pKa = 11.37YY56 pKa = 9.89NSEE59 pKa = 3.84EE60 pKa = 4.06LASLGTNVSISDD72 pKa = 3.53IVIGEE77 pKa = 4.2LGEE80 pKa = 4.73DD81 pKa = 3.57EE82 pKa = 4.65TVADD86 pKa = 3.78IPVGFTLLRR95 pKa = 11.84TAVGAPKK102 pKa = 8.48TTTYY106 pKa = 11.57ADD108 pKa = 3.04TGYY111 pKa = 10.71EE112 pKa = 4.18YY113 pKa = 11.42YY114 pKa = 10.66VLKK117 pKa = 10.71LNQGGGSDD125 pKa = 3.78SPKK128 pKa = 9.08FCDD131 pKa = 3.59RR132 pKa = 11.84IRR134 pKa = 11.84FNYY137 pKa = 10.13EE138 pKa = 3.34GFTLDD143 pKa = 5.45DD144 pKa = 3.62VVFDD148 pKa = 4.19YY149 pKa = 10.97SVHH152 pKa = 6.81PIDD155 pKa = 5.17SDD157 pKa = 4.1LVGDD161 pKa = 4.71GGVTSYY167 pKa = 11.9GLITGWNKK175 pKa = 9.88VMPDD179 pKa = 3.41FNTAEE184 pKa = 4.57GYY186 pKa = 9.84IEE188 pKa = 5.03NGDD191 pKa = 3.68GTVTYY196 pKa = 10.28VNKK199 pKa = 10.52GVGIMFLPSGLAYY212 pKa = 10.45FSSATTGIVSYY223 pKa = 11.0SPIIFKK229 pKa = 10.92FEE231 pKa = 4.2LLQMSQNDD239 pKa = 3.62HH240 pKa = 7.36DD241 pKa = 5.11GDD243 pKa = 4.25GVPSYY248 pKa = 11.42LEE250 pKa = 4.73DD251 pKa = 3.85LPNSNGVRR259 pKa = 11.84DD260 pKa = 3.75SQFTVNYY267 pKa = 10.25DD268 pKa = 3.78DD269 pKa = 5.34LTDD272 pKa = 3.94ATDD275 pKa = 4.96DD276 pKa = 4.03DD277 pKa = 4.58TDD279 pKa = 4.57GDD281 pKa = 4.17GTPDD285 pKa = 4.8YY286 pKa = 10.97IDD288 pKa = 4.15TDD290 pKa = 4.08DD291 pKa = 5.43DD292 pKa = 4.89GDD294 pKa = 4.44GISTINEE301 pKa = 4.68DD302 pKa = 3.78INNDD306 pKa = 3.47GDD308 pKa = 4.04PTNDD312 pKa = 3.51DD313 pKa = 4.02SNGNGIPNYY322 pKa = 10.38LDD324 pKa = 3.03NTDD327 pKa = 3.98IIKK330 pKa = 10.62KK331 pKa = 8.05VTTT334 pKa = 3.81
MM1 pKa = 7.54NLRR4 pKa = 11.84KK5 pKa = 9.85LSLYY9 pKa = 9.92LLCLTIGIVSCKK21 pKa = 10.04KK22 pKa = 10.52DD23 pKa = 3.71DD24 pKa = 4.61DD25 pKa = 4.03PTTVLVEE32 pKa = 3.85IEE34 pKa = 4.47DD35 pKa = 4.57RR36 pKa = 11.84DD37 pKa = 4.02VQQLKK42 pKa = 11.21DD43 pKa = 3.43MDD45 pKa = 4.64SLNKK49 pKa = 10.44YY50 pKa = 10.61LDD52 pKa = 3.23THH54 pKa = 6.69YY55 pKa = 11.37YY56 pKa = 9.89NSEE59 pKa = 3.84EE60 pKa = 4.06LASLGTNVSISDD72 pKa = 3.53IVIGEE77 pKa = 4.2LGEE80 pKa = 4.73DD81 pKa = 3.57EE82 pKa = 4.65TVADD86 pKa = 3.78IPVGFTLLRR95 pKa = 11.84TAVGAPKK102 pKa = 8.48TTTYY106 pKa = 11.57ADD108 pKa = 3.04TGYY111 pKa = 10.71EE112 pKa = 4.18YY113 pKa = 11.42YY114 pKa = 10.66VLKK117 pKa = 10.71LNQGGGSDD125 pKa = 3.78SPKK128 pKa = 9.08FCDD131 pKa = 3.59RR132 pKa = 11.84IRR134 pKa = 11.84FNYY137 pKa = 10.13EE138 pKa = 3.34GFTLDD143 pKa = 5.45DD144 pKa = 3.62VVFDD148 pKa = 4.19YY149 pKa = 10.97SVHH152 pKa = 6.81PIDD155 pKa = 5.17SDD157 pKa = 4.1LVGDD161 pKa = 4.71GGVTSYY167 pKa = 11.9GLITGWNKK175 pKa = 9.88VMPDD179 pKa = 3.41FNTAEE184 pKa = 4.57GYY186 pKa = 9.84IEE188 pKa = 5.03NGDD191 pKa = 3.68GTVTYY196 pKa = 10.28VNKK199 pKa = 10.52GVGIMFLPSGLAYY212 pKa = 10.45FSSATTGIVSYY223 pKa = 11.0SPIIFKK229 pKa = 10.92FEE231 pKa = 4.2LLQMSQNDD239 pKa = 3.62HH240 pKa = 7.36DD241 pKa = 5.11GDD243 pKa = 4.25GVPSYY248 pKa = 11.42LEE250 pKa = 4.73DD251 pKa = 3.85LPNSNGVRR259 pKa = 11.84DD260 pKa = 3.75SQFTVNYY267 pKa = 10.25DD268 pKa = 3.78DD269 pKa = 5.34LTDD272 pKa = 3.94ATDD275 pKa = 4.96DD276 pKa = 4.03DD277 pKa = 4.58TDD279 pKa = 4.57GDD281 pKa = 4.17GTPDD285 pKa = 4.8YY286 pKa = 10.97IDD288 pKa = 4.15TDD290 pKa = 4.08DD291 pKa = 5.43DD292 pKa = 4.89GDD294 pKa = 4.44GISTINEE301 pKa = 4.68DD302 pKa = 3.78INNDD306 pKa = 3.47GDD308 pKa = 4.04PTNDD312 pKa = 3.51DD313 pKa = 4.02SNGNGIPNYY322 pKa = 10.38LDD324 pKa = 3.03NTDD327 pKa = 3.98IIKK330 pKa = 10.62KK331 pKa = 8.05VTTT334 pKa = 3.81
Molecular weight: 36.47 kDa
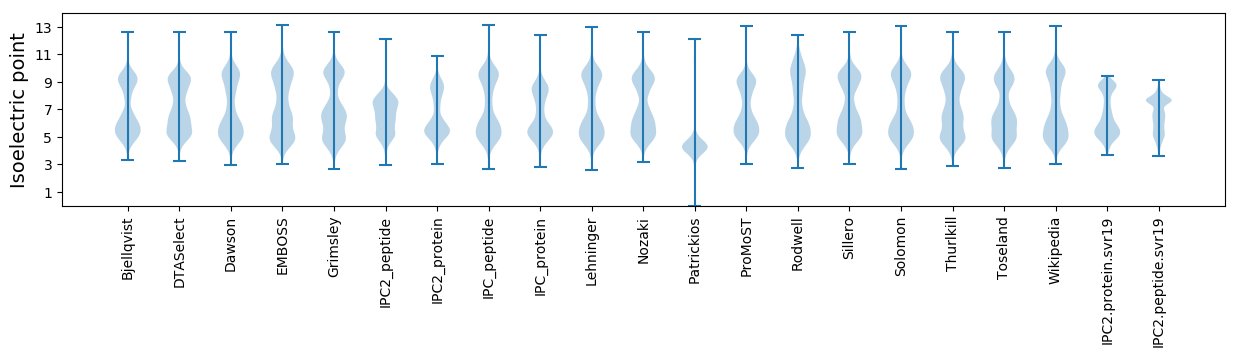
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V2QEQ5|A0A4V2QEQ5_9FLAO TatD DNase family protein OS=Mariniflexile fucanivorans OX=264023 GN=EV196_101374 PE=4 SV=1
MM1 pKa = 7.22KK2 pKa = 10.2RR3 pKa = 11.84ISPRR7 pKa = 11.84MRR9 pKa = 11.84FRR11 pKa = 11.84MMNPFLQFFKK21 pKa = 10.91FIILSIKK28 pKa = 10.26IMVIVAGGHH37 pKa = 5.35GGTRR41 pKa = 11.84KK42 pKa = 10.18VNN44 pKa = 3.26
MM1 pKa = 7.22KK2 pKa = 10.2RR3 pKa = 11.84ISPRR7 pKa = 11.84MRR9 pKa = 11.84FRR11 pKa = 11.84MMNPFLQFFKK21 pKa = 10.91FIILSIKK28 pKa = 10.26IMVIVAGGHH37 pKa = 5.35GGTRR41 pKa = 11.84KK42 pKa = 10.18VNN44 pKa = 3.26
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1414510 |
30 |
4036 |
354.4 |
39.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.164 ± 0.038 | 0.788 ± 0.012 |
5.579 ± 0.029 | 6.335 ± 0.038 |
5.25 ± 0.029 | 6.303 ± 0.039 |
1.767 ± 0.02 | 8.131 ± 0.037 |
7.981 ± 0.055 | 9.152 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.019 | 6.743 ± 0.043 |
3.363 ± 0.022 | 3.215 ± 0.02 |
3.093 ± 0.023 | 6.573 ± 0.033 |
6.075 ± 0.055 | 6.106 ± 0.031 |
1.087 ± 0.014 | 4.165 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
