
Microbulbifer sp. ZGT114
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Microbulbiferaceae; Microbulbifer; unclassified Microbulbifer
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
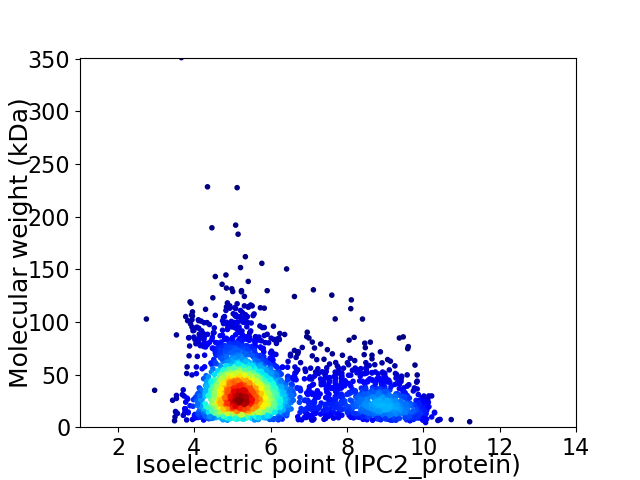
Virtual 2D-PAGE plot for 3054 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
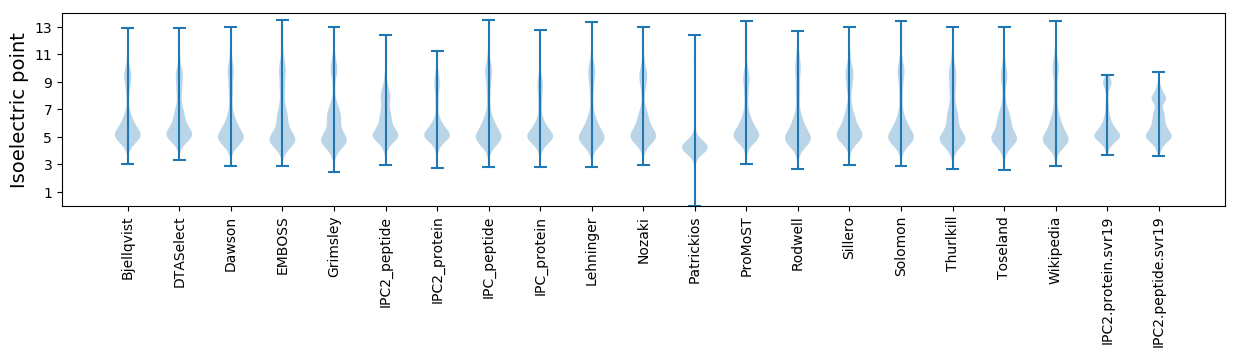
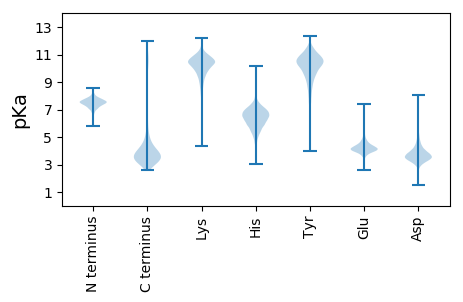
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0X3UCA4|A0A0X3UCA4_9GAMM PDZ domain-containing protein OS=Microbulbifer sp. ZGT114 OX=1685377 GN=AVO43_01180 PE=4 SV=1
MM1 pKa = 7.15KK2 pKa = 10.23HH3 pKa = 6.03VNSKK7 pKa = 10.54LLLSAAIAALLAGCDD22 pKa = 3.19SGGINIEE29 pKa = 4.32PEE31 pKa = 4.29TIDD34 pKa = 3.54NSVDD38 pKa = 3.18NSVVGGGNSEE48 pKa = 4.87DD49 pKa = 3.97NPCASYY55 pKa = 10.7EE56 pKa = 4.03KK57 pKa = 10.41SGQVQQGSFDD67 pKa = 3.9GTNCTYY73 pKa = 10.81AASFVDD79 pKa = 3.69SGNALTQDD87 pKa = 3.82LFIPALEE94 pKa = 4.05NDD96 pKa = 3.75GVHH99 pKa = 6.23IFEE102 pKa = 5.18GSLFVGNNYY111 pKa = 10.6DD112 pKa = 5.03DD113 pKa = 5.07DD114 pKa = 4.16ATMAAAGIAEE124 pKa = 4.7GGDD127 pKa = 3.7GAKK130 pKa = 9.15LTIEE134 pKa = 4.49AGSTIAFPNNTKK146 pKa = 10.57FIVINRR152 pKa = 11.84GSQLFAVGTPDD163 pKa = 3.97APITFTSTSDD173 pKa = 3.79AIDD176 pKa = 3.4NAVAFDD182 pKa = 5.55AVQQWGGMVINGFGITNKK200 pKa = 9.96CSYY203 pKa = 10.28DD204 pKa = 3.55ANLQTSEE211 pKa = 4.17CHH213 pKa = 6.2ILAEE217 pKa = 4.44GAAGKK222 pKa = 9.62DD223 pKa = 3.22QSNYY227 pKa = 10.82GGDD230 pKa = 3.58NNADD234 pKa = 3.12NSGQLEE240 pKa = 4.42YY241 pKa = 11.33VRR243 pKa = 11.84VKK245 pKa = 9.89HH246 pKa = 6.06TGAQVASGDD255 pKa = 4.01EE256 pKa = 4.15LNGITFSSVGSGTIVKK272 pKa = 9.66NLQVYY277 pKa = 7.67STYY280 pKa = 11.19DD281 pKa = 3.1DD282 pKa = 4.67GIEE285 pKa = 4.02MFGGAVSFEE294 pKa = 4.78NYY296 pKa = 9.33LAMYY300 pKa = 10.03VRR302 pKa = 11.84DD303 pKa = 4.14DD304 pKa = 5.1SIDD307 pKa = 3.38IDD309 pKa = 4.09EE310 pKa = 4.88GWAGSITNALVIQSEE325 pKa = 4.59TDD327 pKa = 3.22GNHH330 pKa = 6.72CIEE333 pKa = 4.25SDD335 pKa = 4.26GIGSYY340 pKa = 10.63DD341 pKa = 3.89DD342 pKa = 4.21KK343 pKa = 11.53DD344 pKa = 3.34AAFIDD349 pKa = 4.59GVISQGINSRR359 pKa = 11.84PVIDD363 pKa = 5.01GLTCIISANGADD375 pKa = 4.2TATHH379 pKa = 6.97DD380 pKa = 4.54PGAGWRR386 pKa = 11.84FRR388 pKa = 11.84EE389 pKa = 4.63AIFPKK394 pKa = 8.92ITNSMVVGSFSADD407 pKa = 3.1TTGVDD412 pKa = 3.35GSNYY416 pKa = 9.15CLRR419 pKa = 11.84VEE421 pKa = 4.44DD422 pKa = 4.89PEE424 pKa = 4.44TLVEE428 pKa = 4.25AEE430 pKa = 4.74VGTEE434 pKa = 3.83LQIEE438 pKa = 4.58SNIFACEE445 pKa = 3.87DD446 pKa = 3.43RR447 pKa = 11.84TKK449 pKa = 10.7GGPVGTQTLEE459 pKa = 3.49EE460 pKa = 3.99WAVANGNVFADD471 pKa = 3.27ITGAVDD477 pKa = 3.59PTAMANTEE485 pKa = 4.04LQILEE490 pKa = 4.25GVPSVFSIDD499 pKa = 3.47AASMQVNGAAIGISPIEE516 pKa = 3.75RR517 pKa = 11.84AYY519 pKa = 10.77FGALSQGDD527 pKa = 4.51LDD529 pKa = 3.86WTLGWTYY536 pKa = 11.21GLHH539 pKa = 6.04NGEE542 pKa = 4.94ALWFANN548 pKa = 3.92
MM1 pKa = 7.15KK2 pKa = 10.23HH3 pKa = 6.03VNSKK7 pKa = 10.54LLLSAAIAALLAGCDD22 pKa = 3.19SGGINIEE29 pKa = 4.32PEE31 pKa = 4.29TIDD34 pKa = 3.54NSVDD38 pKa = 3.18NSVVGGGNSEE48 pKa = 4.87DD49 pKa = 3.97NPCASYY55 pKa = 10.7EE56 pKa = 4.03KK57 pKa = 10.41SGQVQQGSFDD67 pKa = 3.9GTNCTYY73 pKa = 10.81AASFVDD79 pKa = 3.69SGNALTQDD87 pKa = 3.82LFIPALEE94 pKa = 4.05NDD96 pKa = 3.75GVHH99 pKa = 6.23IFEE102 pKa = 5.18GSLFVGNNYY111 pKa = 10.6DD112 pKa = 5.03DD113 pKa = 5.07DD114 pKa = 4.16ATMAAAGIAEE124 pKa = 4.7GGDD127 pKa = 3.7GAKK130 pKa = 9.15LTIEE134 pKa = 4.49AGSTIAFPNNTKK146 pKa = 10.57FIVINRR152 pKa = 11.84GSQLFAVGTPDD163 pKa = 3.97APITFTSTSDD173 pKa = 3.79AIDD176 pKa = 3.4NAVAFDD182 pKa = 5.55AVQQWGGMVINGFGITNKK200 pKa = 9.96CSYY203 pKa = 10.28DD204 pKa = 3.55ANLQTSEE211 pKa = 4.17CHH213 pKa = 6.2ILAEE217 pKa = 4.44GAAGKK222 pKa = 9.62DD223 pKa = 3.22QSNYY227 pKa = 10.82GGDD230 pKa = 3.58NNADD234 pKa = 3.12NSGQLEE240 pKa = 4.42YY241 pKa = 11.33VRR243 pKa = 11.84VKK245 pKa = 9.89HH246 pKa = 6.06TGAQVASGDD255 pKa = 4.01EE256 pKa = 4.15LNGITFSSVGSGTIVKK272 pKa = 9.66NLQVYY277 pKa = 7.67STYY280 pKa = 11.19DD281 pKa = 3.1DD282 pKa = 4.67GIEE285 pKa = 4.02MFGGAVSFEE294 pKa = 4.78NYY296 pKa = 9.33LAMYY300 pKa = 10.03VRR302 pKa = 11.84DD303 pKa = 4.14DD304 pKa = 5.1SIDD307 pKa = 3.38IDD309 pKa = 4.09EE310 pKa = 4.88GWAGSITNALVIQSEE325 pKa = 4.59TDD327 pKa = 3.22GNHH330 pKa = 6.72CIEE333 pKa = 4.25SDD335 pKa = 4.26GIGSYY340 pKa = 10.63DD341 pKa = 3.89DD342 pKa = 4.21KK343 pKa = 11.53DD344 pKa = 3.34AAFIDD349 pKa = 4.59GVISQGINSRR359 pKa = 11.84PVIDD363 pKa = 5.01GLTCIISANGADD375 pKa = 4.2TATHH379 pKa = 6.97DD380 pKa = 4.54PGAGWRR386 pKa = 11.84FRR388 pKa = 11.84EE389 pKa = 4.63AIFPKK394 pKa = 8.92ITNSMVVGSFSADD407 pKa = 3.1TTGVDD412 pKa = 3.35GSNYY416 pKa = 9.15CLRR419 pKa = 11.84VEE421 pKa = 4.44DD422 pKa = 4.89PEE424 pKa = 4.44TLVEE428 pKa = 4.25AEE430 pKa = 4.74VGTEE434 pKa = 3.83LQIEE438 pKa = 4.58SNIFACEE445 pKa = 3.87DD446 pKa = 3.43RR447 pKa = 11.84TKK449 pKa = 10.7GGPVGTQTLEE459 pKa = 3.49EE460 pKa = 3.99WAVANGNVFADD471 pKa = 3.27ITGAVDD477 pKa = 3.59PTAMANTEE485 pKa = 4.04LQILEE490 pKa = 4.25GVPSVFSIDD499 pKa = 3.47AASMQVNGAAIGISPIEE516 pKa = 3.75RR517 pKa = 11.84AYY519 pKa = 10.77FGALSQGDD527 pKa = 4.51LDD529 pKa = 3.86WTLGWTYY536 pKa = 11.21GLHH539 pKa = 6.04NGEE542 pKa = 4.94ALWFANN548 pKa = 3.92
Molecular weight: 57.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X3U4Z2|A0A0X3U4Z2_9GAMM Gamma carbonic anhydrase family protein OS=Microbulbifer sp. ZGT114 OX=1685377 GN=AVO43_11040 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.16RR12 pKa = 11.84KK13 pKa = 9.08RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.87ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.16RR12 pKa = 11.84KK13 pKa = 9.08RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 8.87ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1034147 |
38 |
3367 |
338.6 |
37.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.498 ± 0.051 | 0.991 ± 0.017 |
5.733 ± 0.038 | 6.894 ± 0.043 |
3.754 ± 0.027 | 8.048 ± 0.037 |
2.13 ± 0.024 | 4.846 ± 0.032 |
3.467 ± 0.037 | 10.794 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.302 ± 0.021 | 3.088 ± 0.029 |
4.72 ± 0.029 | 4.175 ± 0.032 |
6.697 ± 0.047 | 5.824 ± 0.03 |
4.807 ± 0.037 | 7.087 ± 0.043 |
1.404 ± 0.019 | 2.741 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
