
Wenling narna-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.31
Get precalculated fractions of proteins
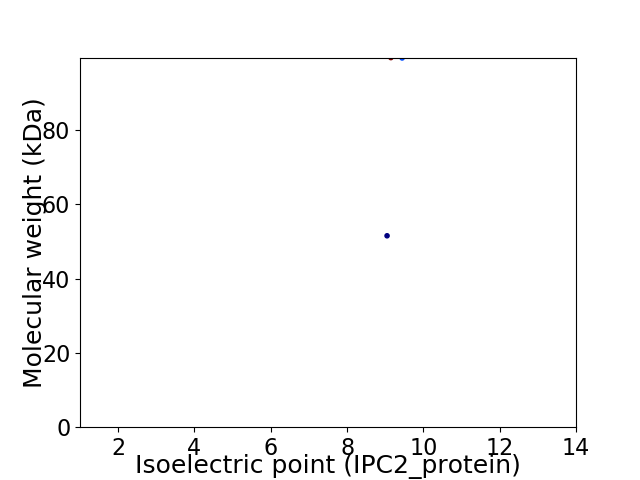
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIV3|A0A1L3KIV3_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 7 OX=1923507 PE=4 SV=1
MM1 pKa = 6.2VTHH4 pKa = 7.15HH5 pKa = 6.66ARR7 pKa = 11.84EE8 pKa = 4.0EE9 pKa = 3.88LAQGVGHH16 pKa = 6.91PPRR19 pKa = 11.84CEE21 pKa = 4.1SEE23 pKa = 5.14LSLQRR28 pKa = 11.84PFLGPEE34 pKa = 3.46EE35 pKa = 4.67DD36 pKa = 3.6AQFTLKK42 pKa = 10.35FHH44 pKa = 7.3RR45 pKa = 11.84EE46 pKa = 4.04VLDD49 pKa = 4.06SFQTSQKK56 pKa = 10.19VKK58 pKa = 10.1PGWRR62 pKa = 11.84CAPLADD68 pKa = 4.06RR69 pKa = 11.84EE70 pKa = 4.44GRR72 pKa = 11.84RR73 pKa = 11.84GNPDD77 pKa = 2.64VRR79 pKa = 11.84VFSGHH84 pKa = 5.89SLPGYY89 pKa = 9.83RR90 pKa = 11.84PADD93 pKa = 3.84PFPAARR99 pKa = 11.84GPLEE103 pKa = 4.05VLPLAVQKK111 pKa = 9.41PCCEE115 pKa = 4.03AVSQPLGQRR124 pKa = 11.84HH125 pKa = 5.27AAHH128 pKa = 7.14RR129 pKa = 11.84SSQRR133 pKa = 11.84RR134 pKa = 11.84QAISPDD140 pKa = 3.39YY141 pKa = 9.72QRR143 pKa = 11.84QGRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.89SSLPRR153 pKa = 11.84PAPNARR159 pKa = 11.84VEE161 pKa = 4.11DD162 pKa = 3.74AKK164 pKa = 11.17YY165 pKa = 10.79SPNSHH170 pKa = 6.69RR171 pKa = 11.84GAKK174 pKa = 9.14IHH176 pKa = 6.9GSLHH180 pKa = 6.4RR181 pKa = 11.84CRR183 pKa = 11.84QRR185 pKa = 11.84SPGRR189 pKa = 11.84SKK191 pKa = 10.84AAILGRR197 pKa = 11.84VKK199 pKa = 10.75RR200 pKa = 11.84SDD202 pKa = 3.73QPPKK206 pKa = 10.73GYY208 pKa = 10.41AADD211 pKa = 4.04GGEE214 pKa = 4.3LCLNPIPLAHH224 pKa = 6.47GRR226 pKa = 11.84LPRR229 pKa = 11.84RR230 pKa = 11.84PGNRR234 pKa = 11.84EE235 pKa = 3.84EE236 pKa = 4.06VCEE239 pKa = 3.97RR240 pKa = 11.84CSPRR244 pKa = 11.84FRR246 pKa = 11.84RR247 pKa = 11.84IHH249 pKa = 4.94EE250 pKa = 4.46TVFLLLPRR258 pKa = 11.84DD259 pKa = 3.64LKK261 pKa = 11.39DD262 pKa = 4.12LFGEE266 pKa = 4.3KK267 pKa = 10.21NVVSGSVMFTDD278 pKa = 4.35RR279 pKa = 11.84EE280 pKa = 4.06LRR282 pKa = 11.84SAGFRR287 pKa = 11.84HH288 pKa = 5.79FLVSVGHH295 pKa = 6.08FWWPDD300 pKa = 2.82RR301 pKa = 11.84RR302 pKa = 11.84KK303 pKa = 10.29VVATDD308 pKa = 3.33DD309 pKa = 3.73RR310 pKa = 11.84PGRR313 pKa = 11.84RR314 pKa = 11.84CRR316 pKa = 11.84LGGQGNVLEE325 pKa = 4.94GEE327 pKa = 4.38TPRR330 pKa = 11.84RR331 pKa = 11.84GPGRR335 pKa = 11.84LGPKK339 pKa = 8.88EE340 pKa = 3.95VNEE343 pKa = 4.18IKK345 pKa = 10.36QGPRR349 pKa = 11.84RR350 pKa = 11.84RR351 pKa = 11.84EE352 pKa = 3.8AHH354 pKa = 6.87HH355 pKa = 7.23DD356 pKa = 3.69PPPDD360 pKa = 3.78RR361 pKa = 11.84EE362 pKa = 4.31TVAIRR367 pKa = 11.84QTSIQGTCQFSEE379 pKa = 4.68DD380 pKa = 4.21LLNLCRR386 pKa = 11.84WRR388 pKa = 11.84LSRR391 pKa = 11.84TGEE394 pKa = 3.8EE395 pKa = 4.38TIDD398 pKa = 3.4KK399 pKa = 10.99GRR401 pKa = 11.84DD402 pKa = 3.2QVLRR406 pKa = 11.84EE407 pKa = 4.07QICSGSQVGGEE418 pKa = 3.92ADD420 pKa = 3.56VLLLRR425 pKa = 11.84RR426 pKa = 11.84LLTVNPLGLSPEE438 pKa = 4.34GPRR441 pKa = 11.84VIPRR445 pKa = 11.84QDD447 pKa = 3.23ALHH450 pKa = 6.65APVFPEE456 pKa = 3.86TTEE459 pKa = 3.76
MM1 pKa = 6.2VTHH4 pKa = 7.15HH5 pKa = 6.66ARR7 pKa = 11.84EE8 pKa = 4.0EE9 pKa = 3.88LAQGVGHH16 pKa = 6.91PPRR19 pKa = 11.84CEE21 pKa = 4.1SEE23 pKa = 5.14LSLQRR28 pKa = 11.84PFLGPEE34 pKa = 3.46EE35 pKa = 4.67DD36 pKa = 3.6AQFTLKK42 pKa = 10.35FHH44 pKa = 7.3RR45 pKa = 11.84EE46 pKa = 4.04VLDD49 pKa = 4.06SFQTSQKK56 pKa = 10.19VKK58 pKa = 10.1PGWRR62 pKa = 11.84CAPLADD68 pKa = 4.06RR69 pKa = 11.84EE70 pKa = 4.44GRR72 pKa = 11.84RR73 pKa = 11.84GNPDD77 pKa = 2.64VRR79 pKa = 11.84VFSGHH84 pKa = 5.89SLPGYY89 pKa = 9.83RR90 pKa = 11.84PADD93 pKa = 3.84PFPAARR99 pKa = 11.84GPLEE103 pKa = 4.05VLPLAVQKK111 pKa = 9.41PCCEE115 pKa = 4.03AVSQPLGQRR124 pKa = 11.84HH125 pKa = 5.27AAHH128 pKa = 7.14RR129 pKa = 11.84SSQRR133 pKa = 11.84RR134 pKa = 11.84QAISPDD140 pKa = 3.39YY141 pKa = 9.72QRR143 pKa = 11.84QGRR146 pKa = 11.84RR147 pKa = 11.84EE148 pKa = 3.89SSLPRR153 pKa = 11.84PAPNARR159 pKa = 11.84VEE161 pKa = 4.11DD162 pKa = 3.74AKK164 pKa = 11.17YY165 pKa = 10.79SPNSHH170 pKa = 6.69RR171 pKa = 11.84GAKK174 pKa = 9.14IHH176 pKa = 6.9GSLHH180 pKa = 6.4RR181 pKa = 11.84CRR183 pKa = 11.84QRR185 pKa = 11.84SPGRR189 pKa = 11.84SKK191 pKa = 10.84AAILGRR197 pKa = 11.84VKK199 pKa = 10.75RR200 pKa = 11.84SDD202 pKa = 3.73QPPKK206 pKa = 10.73GYY208 pKa = 10.41AADD211 pKa = 4.04GGEE214 pKa = 4.3LCLNPIPLAHH224 pKa = 6.47GRR226 pKa = 11.84LPRR229 pKa = 11.84RR230 pKa = 11.84PGNRR234 pKa = 11.84EE235 pKa = 3.84EE236 pKa = 4.06VCEE239 pKa = 3.97RR240 pKa = 11.84CSPRR244 pKa = 11.84FRR246 pKa = 11.84RR247 pKa = 11.84IHH249 pKa = 4.94EE250 pKa = 4.46TVFLLLPRR258 pKa = 11.84DD259 pKa = 3.64LKK261 pKa = 11.39DD262 pKa = 4.12LFGEE266 pKa = 4.3KK267 pKa = 10.21NVVSGSVMFTDD278 pKa = 4.35RR279 pKa = 11.84EE280 pKa = 4.06LRR282 pKa = 11.84SAGFRR287 pKa = 11.84HH288 pKa = 5.79FLVSVGHH295 pKa = 6.08FWWPDD300 pKa = 2.82RR301 pKa = 11.84RR302 pKa = 11.84KK303 pKa = 10.29VVATDD308 pKa = 3.33DD309 pKa = 3.73RR310 pKa = 11.84PGRR313 pKa = 11.84RR314 pKa = 11.84CRR316 pKa = 11.84LGGQGNVLEE325 pKa = 4.94GEE327 pKa = 4.38TPRR330 pKa = 11.84RR331 pKa = 11.84GPGRR335 pKa = 11.84LGPKK339 pKa = 8.88EE340 pKa = 3.95VNEE343 pKa = 4.18IKK345 pKa = 10.36QGPRR349 pKa = 11.84RR350 pKa = 11.84RR351 pKa = 11.84EE352 pKa = 3.8AHH354 pKa = 6.87HH355 pKa = 7.23DD356 pKa = 3.69PPPDD360 pKa = 3.78RR361 pKa = 11.84EE362 pKa = 4.31TVAIRR367 pKa = 11.84QTSIQGTCQFSEE379 pKa = 4.68DD380 pKa = 4.21LLNLCRR386 pKa = 11.84WRR388 pKa = 11.84LSRR391 pKa = 11.84TGEE394 pKa = 3.8EE395 pKa = 4.38TIDD398 pKa = 3.4KK399 pKa = 10.99GRR401 pKa = 11.84DD402 pKa = 3.2QVLRR406 pKa = 11.84EE407 pKa = 4.07QICSGSQVGGEE418 pKa = 3.92ADD420 pKa = 3.56VLLLRR425 pKa = 11.84RR426 pKa = 11.84LLTVNPLGLSPEE438 pKa = 4.34GPRR441 pKa = 11.84VIPRR445 pKa = 11.84QDD447 pKa = 3.23ALHH450 pKa = 6.65APVFPEE456 pKa = 3.86TTEE459 pKa = 3.76
Molecular weight: 51.53 kDa
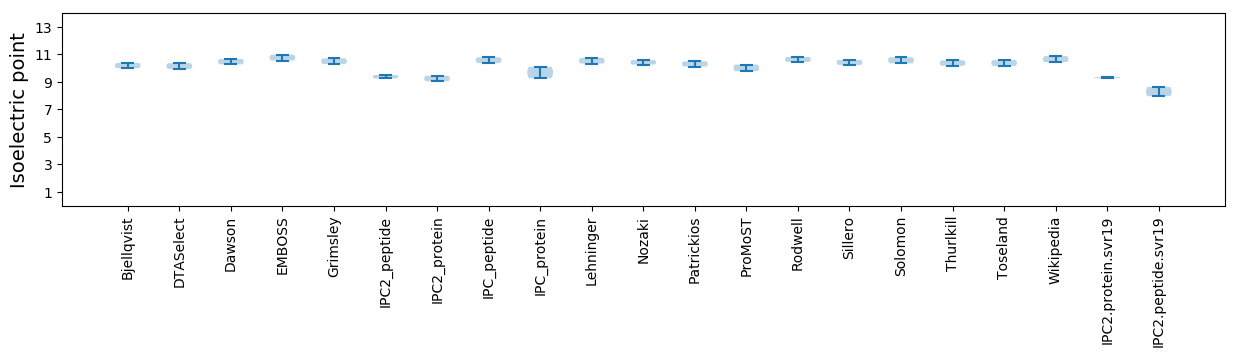
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIV3|A0A1L3KIV3_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 7 OX=1923507 PE=4 SV=1
MM1 pKa = 7.57PRR3 pKa = 11.84NKK5 pKa = 9.75ARR7 pKa = 11.84HH8 pKa = 5.33RR9 pKa = 11.84WVCLYY14 pKa = 10.71EE15 pKa = 4.16LTLSVATLLPARR27 pKa = 11.84PGTGPYY33 pKa = 10.37LGGTRR38 pKa = 11.84PNGPDD43 pKa = 2.73TGRR46 pKa = 11.84FLPRR50 pKa = 11.84FPRR53 pKa = 11.84HH54 pKa = 5.44PNIRR58 pKa = 11.84CPKK61 pKa = 8.44EE62 pKa = 3.67RR63 pKa = 11.84GNAATGKK70 pKa = 5.35TTNGNVNGILSKK82 pKa = 9.92VLRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PLGNQRR93 pKa = 11.84WTRR96 pKa = 11.84LEE98 pKa = 4.04EE99 pKa = 3.55AWRR102 pKa = 11.84TILAAIAACGFVRR115 pKa = 11.84LDD117 pKa = 3.3SSRR120 pKa = 11.84RR121 pKa = 11.84AHH123 pKa = 6.93RR124 pKa = 11.84EE125 pKa = 3.37ALRR128 pKa = 11.84GLSRR132 pKa = 11.84LSTWIVKK139 pKa = 8.15TAALSGTEE147 pKa = 3.85EE148 pKa = 4.08VLRR151 pKa = 11.84EE152 pKa = 4.06LKK154 pKa = 9.04TWSARR159 pKa = 11.84LRR161 pKa = 11.84AHH163 pKa = 6.22VVNSLPPHH171 pKa = 5.77KK172 pKa = 9.81RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84GFARR179 pKa = 11.84YY180 pKa = 7.98FQGVLRR186 pKa = 11.84SLIDD190 pKa = 3.36KK191 pKa = 10.42DD192 pKa = 3.95SRR194 pKa = 11.84ALQMSYY200 pKa = 10.01IGRR203 pKa = 11.84ALPVGSFRR211 pKa = 11.84EE212 pKa = 4.12TEE214 pKa = 3.75GAIRR218 pKa = 11.84KK219 pKa = 8.55FRR221 pKa = 11.84EE222 pKa = 4.01VTQVPFEE229 pKa = 4.1THH231 pKa = 7.16PDD233 pKa = 3.48LLDD236 pKa = 4.47SIRR239 pKa = 11.84DD240 pKa = 3.73WGAHH244 pKa = 3.75WAARR248 pKa = 11.84FLDD251 pKa = 4.48TPAMPPLWRR260 pKa = 11.84SSSGCVDD267 pKa = 4.23FSRR270 pKa = 11.84LQGGLAEE277 pKa = 4.33AAVRR281 pKa = 11.84AADD284 pKa = 4.26FAAAEE289 pKa = 4.3VEE291 pKa = 4.97SRR293 pKa = 11.84EE294 pKa = 4.29DD295 pKa = 3.2WDD297 pKa = 4.53DD298 pKa = 3.74LRR300 pKa = 11.84NTAAEE305 pKa = 4.28VPEE308 pKa = 4.18MTDD311 pKa = 3.06CDD313 pKa = 3.1VDD315 pKa = 3.49IAVRR319 pKa = 11.84EE320 pKa = 4.05AQLRR324 pKa = 11.84DD325 pKa = 3.34VALTVAAHH333 pKa = 6.66LPDD336 pKa = 4.66LPPVEE341 pKa = 4.5VCTVPEE347 pKa = 4.05RR348 pKa = 11.84GMKK351 pKa = 9.98CRR353 pKa = 11.84IVTKK357 pKa = 10.61APWCLIHH364 pKa = 7.1IGHH367 pKa = 7.48FLRR370 pKa = 11.84SWLFGGLRR378 pKa = 11.84KK379 pKa = 9.76DD380 pKa = 3.34RR381 pKa = 11.84RR382 pKa = 11.84VQGVLAGDD390 pKa = 3.36HH391 pKa = 6.68AGALRR396 pKa = 11.84GQTEE400 pKa = 4.01RR401 pKa = 11.84VYY403 pKa = 10.44SQKK406 pKa = 9.2PTEE409 pKa = 4.32EE410 pKa = 4.03EE411 pKa = 4.23NICLSADD418 pKa = 3.38LTAATDD424 pKa = 4.46LFPQDD429 pKa = 3.66LVAALVDD436 pKa = 4.01GLLSGARR443 pKa = 11.84KK444 pKa = 8.99PPTAEE449 pKa = 3.48IQEE452 pKa = 4.4VFRR455 pKa = 11.84KK456 pKa = 8.53LTGPLNARR464 pKa = 11.84LPDD467 pKa = 3.84GDD469 pKa = 3.64GFAIRR474 pKa = 11.84RR475 pKa = 11.84GIMMGFPTTWTLLNLVNLFWSEE497 pKa = 4.37TAWSASGSLPFQDD510 pKa = 3.8VALSAQPAASPRR522 pKa = 11.84TIVCGDD528 pKa = 3.87DD529 pKa = 4.06LASVWPPKK537 pKa = 10.3VADD540 pKa = 4.4RR541 pKa = 11.84YY542 pKa = 10.33EE543 pKa = 4.27EE544 pKa = 3.99VAEE547 pKa = 4.06ACGAKK552 pKa = 10.26FSVGKK557 pKa = 9.87HH558 pKa = 4.02YY559 pKa = 10.69RR560 pKa = 11.84SRR562 pKa = 11.84DD563 pKa = 3.49YY564 pKa = 11.22ILFTEE569 pKa = 5.21EE570 pKa = 3.45IFKK573 pKa = 10.97VSWEE577 pKa = 4.02KK578 pKa = 10.89QEE580 pKa = 3.91YY581 pKa = 10.93SLMDD585 pKa = 3.35SAKK588 pKa = 10.03PRR590 pKa = 11.84RR591 pKa = 11.84TTLADD596 pKa = 4.08FFPVPRR602 pKa = 11.84APRR605 pKa = 11.84KK606 pKa = 7.65STMRR610 pKa = 11.84KK611 pKa = 9.22RR612 pKa = 11.84YY613 pKa = 9.13RR614 pKa = 11.84VQAKK618 pKa = 8.7LAPVRR623 pKa = 11.84CVPLRR628 pKa = 11.84GLVRR632 pKa = 11.84PLHH635 pKa = 5.74APKK638 pKa = 10.29DD639 pKa = 3.68RR640 pKa = 11.84RR641 pKa = 11.84LRR643 pKa = 11.84PSWAALPAAVEE654 pKa = 4.01AAMDD658 pKa = 4.28LGAPVAVRR666 pKa = 11.84RR667 pKa = 11.84VLRR670 pKa = 11.84VLHH673 pKa = 5.96PGIWCWARR681 pKa = 11.84KK682 pKa = 9.5RR683 pKa = 11.84GFSPTLPLVVGGYY696 pKa = 9.83GLPPLRR702 pKa = 11.84GSVRR706 pKa = 11.84RR707 pKa = 11.84VSLPKK712 pKa = 8.97WLRR715 pKa = 11.84YY716 pKa = 9.37GLAAWLLNSKK726 pKa = 9.32WKK728 pKa = 10.43DD729 pKa = 3.2LEE731 pKa = 4.56GPSRR735 pKa = 11.84CWEE738 pKa = 4.41GVCRR742 pKa = 11.84PVSWQTMARR751 pKa = 11.84EE752 pKa = 4.04HH753 pKa = 6.7ADD755 pKa = 3.18VRR757 pKa = 11.84IASSTFAVRR766 pKa = 11.84KK767 pKa = 9.88GRR769 pKa = 11.84TPPPGFHH776 pKa = 7.12LLGGLEE782 pKa = 4.64AIQNLSVKK790 pKa = 10.38FEE792 pKa = 4.36GEE794 pKa = 4.01LCVLLGTEE802 pKa = 4.16EE803 pKa = 4.68GSLKK807 pKa = 10.47RR808 pKa = 11.84QFGLAPRR815 pKa = 11.84RR816 pKa = 11.84VANSLRR822 pKa = 11.84KK823 pKa = 9.23FFARR827 pKa = 11.84MVSDD831 pKa = 3.54HH832 pKa = 7.0PSRR835 pKa = 11.84RR836 pKa = 11.84PFPSSLGRR844 pKa = 11.84TALVQRR850 pKa = 11.84WRR852 pKa = 11.84SRR854 pKa = 11.84AEE856 pKa = 3.48EE857 pKa = 3.85RR858 pKa = 11.84ALYY861 pKa = 7.48STPRR865 pKa = 11.84SAVEE869 pKa = 3.89GLVLSLPVRR878 pKa = 11.84AKK880 pKa = 10.59RR881 pKa = 11.84LLAEE885 pKa = 4.09ALGWVV890 pKa = 3.71
MM1 pKa = 7.57PRR3 pKa = 11.84NKK5 pKa = 9.75ARR7 pKa = 11.84HH8 pKa = 5.33RR9 pKa = 11.84WVCLYY14 pKa = 10.71EE15 pKa = 4.16LTLSVATLLPARR27 pKa = 11.84PGTGPYY33 pKa = 10.37LGGTRR38 pKa = 11.84PNGPDD43 pKa = 2.73TGRR46 pKa = 11.84FLPRR50 pKa = 11.84FPRR53 pKa = 11.84HH54 pKa = 5.44PNIRR58 pKa = 11.84CPKK61 pKa = 8.44EE62 pKa = 3.67RR63 pKa = 11.84GNAATGKK70 pKa = 5.35TTNGNVNGILSKK82 pKa = 9.92VLRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PLGNQRR93 pKa = 11.84WTRR96 pKa = 11.84LEE98 pKa = 4.04EE99 pKa = 3.55AWRR102 pKa = 11.84TILAAIAACGFVRR115 pKa = 11.84LDD117 pKa = 3.3SSRR120 pKa = 11.84RR121 pKa = 11.84AHH123 pKa = 6.93RR124 pKa = 11.84EE125 pKa = 3.37ALRR128 pKa = 11.84GLSRR132 pKa = 11.84LSTWIVKK139 pKa = 8.15TAALSGTEE147 pKa = 3.85EE148 pKa = 4.08VLRR151 pKa = 11.84EE152 pKa = 4.06LKK154 pKa = 9.04TWSARR159 pKa = 11.84LRR161 pKa = 11.84AHH163 pKa = 6.22VVNSLPPHH171 pKa = 5.77KK172 pKa = 9.81RR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84GFARR179 pKa = 11.84YY180 pKa = 7.98FQGVLRR186 pKa = 11.84SLIDD190 pKa = 3.36KK191 pKa = 10.42DD192 pKa = 3.95SRR194 pKa = 11.84ALQMSYY200 pKa = 10.01IGRR203 pKa = 11.84ALPVGSFRR211 pKa = 11.84EE212 pKa = 4.12TEE214 pKa = 3.75GAIRR218 pKa = 11.84KK219 pKa = 8.55FRR221 pKa = 11.84EE222 pKa = 4.01VTQVPFEE229 pKa = 4.1THH231 pKa = 7.16PDD233 pKa = 3.48LLDD236 pKa = 4.47SIRR239 pKa = 11.84DD240 pKa = 3.73WGAHH244 pKa = 3.75WAARR248 pKa = 11.84FLDD251 pKa = 4.48TPAMPPLWRR260 pKa = 11.84SSSGCVDD267 pKa = 4.23FSRR270 pKa = 11.84LQGGLAEE277 pKa = 4.33AAVRR281 pKa = 11.84AADD284 pKa = 4.26FAAAEE289 pKa = 4.3VEE291 pKa = 4.97SRR293 pKa = 11.84EE294 pKa = 4.29DD295 pKa = 3.2WDD297 pKa = 4.53DD298 pKa = 3.74LRR300 pKa = 11.84NTAAEE305 pKa = 4.28VPEE308 pKa = 4.18MTDD311 pKa = 3.06CDD313 pKa = 3.1VDD315 pKa = 3.49IAVRR319 pKa = 11.84EE320 pKa = 4.05AQLRR324 pKa = 11.84DD325 pKa = 3.34VALTVAAHH333 pKa = 6.66LPDD336 pKa = 4.66LPPVEE341 pKa = 4.5VCTVPEE347 pKa = 4.05RR348 pKa = 11.84GMKK351 pKa = 9.98CRR353 pKa = 11.84IVTKK357 pKa = 10.61APWCLIHH364 pKa = 7.1IGHH367 pKa = 7.48FLRR370 pKa = 11.84SWLFGGLRR378 pKa = 11.84KK379 pKa = 9.76DD380 pKa = 3.34RR381 pKa = 11.84RR382 pKa = 11.84VQGVLAGDD390 pKa = 3.36HH391 pKa = 6.68AGALRR396 pKa = 11.84GQTEE400 pKa = 4.01RR401 pKa = 11.84VYY403 pKa = 10.44SQKK406 pKa = 9.2PTEE409 pKa = 4.32EE410 pKa = 4.03EE411 pKa = 4.23NICLSADD418 pKa = 3.38LTAATDD424 pKa = 4.46LFPQDD429 pKa = 3.66LVAALVDD436 pKa = 4.01GLLSGARR443 pKa = 11.84KK444 pKa = 8.99PPTAEE449 pKa = 3.48IQEE452 pKa = 4.4VFRR455 pKa = 11.84KK456 pKa = 8.53LTGPLNARR464 pKa = 11.84LPDD467 pKa = 3.84GDD469 pKa = 3.64GFAIRR474 pKa = 11.84RR475 pKa = 11.84GIMMGFPTTWTLLNLVNLFWSEE497 pKa = 4.37TAWSASGSLPFQDD510 pKa = 3.8VALSAQPAASPRR522 pKa = 11.84TIVCGDD528 pKa = 3.87DD529 pKa = 4.06LASVWPPKK537 pKa = 10.3VADD540 pKa = 4.4RR541 pKa = 11.84YY542 pKa = 10.33EE543 pKa = 4.27EE544 pKa = 3.99VAEE547 pKa = 4.06ACGAKK552 pKa = 10.26FSVGKK557 pKa = 9.87HH558 pKa = 4.02YY559 pKa = 10.69RR560 pKa = 11.84SRR562 pKa = 11.84DD563 pKa = 3.49YY564 pKa = 11.22ILFTEE569 pKa = 5.21EE570 pKa = 3.45IFKK573 pKa = 10.97VSWEE577 pKa = 4.02KK578 pKa = 10.89QEE580 pKa = 3.91YY581 pKa = 10.93SLMDD585 pKa = 3.35SAKK588 pKa = 10.03PRR590 pKa = 11.84RR591 pKa = 11.84TTLADD596 pKa = 4.08FFPVPRR602 pKa = 11.84APRR605 pKa = 11.84KK606 pKa = 7.65STMRR610 pKa = 11.84KK611 pKa = 9.22RR612 pKa = 11.84YY613 pKa = 9.13RR614 pKa = 11.84VQAKK618 pKa = 8.7LAPVRR623 pKa = 11.84CVPLRR628 pKa = 11.84GLVRR632 pKa = 11.84PLHH635 pKa = 5.74APKK638 pKa = 10.29DD639 pKa = 3.68RR640 pKa = 11.84RR641 pKa = 11.84LRR643 pKa = 11.84PSWAALPAAVEE654 pKa = 4.01AAMDD658 pKa = 4.28LGAPVAVRR666 pKa = 11.84RR667 pKa = 11.84VLRR670 pKa = 11.84VLHH673 pKa = 5.96PGIWCWARR681 pKa = 11.84KK682 pKa = 9.5RR683 pKa = 11.84GFSPTLPLVVGGYY696 pKa = 9.83GLPPLRR702 pKa = 11.84GSVRR706 pKa = 11.84RR707 pKa = 11.84VSLPKK712 pKa = 8.97WLRR715 pKa = 11.84YY716 pKa = 9.37GLAAWLLNSKK726 pKa = 9.32WKK728 pKa = 10.43DD729 pKa = 3.2LEE731 pKa = 4.56GPSRR735 pKa = 11.84CWEE738 pKa = 4.41GVCRR742 pKa = 11.84PVSWQTMARR751 pKa = 11.84EE752 pKa = 4.04HH753 pKa = 6.7ADD755 pKa = 3.18VRR757 pKa = 11.84IASSTFAVRR766 pKa = 11.84KK767 pKa = 9.88GRR769 pKa = 11.84TPPPGFHH776 pKa = 7.12LLGGLEE782 pKa = 4.64AIQNLSVKK790 pKa = 10.38FEE792 pKa = 4.36GEE794 pKa = 4.01LCVLLGTEE802 pKa = 4.16EE803 pKa = 4.68GSLKK807 pKa = 10.47RR808 pKa = 11.84QFGLAPRR815 pKa = 11.84RR816 pKa = 11.84VANSLRR822 pKa = 11.84KK823 pKa = 9.23FFARR827 pKa = 11.84MVSDD831 pKa = 3.54HH832 pKa = 7.0PSRR835 pKa = 11.84RR836 pKa = 11.84PFPSSLGRR844 pKa = 11.84TALVQRR850 pKa = 11.84WRR852 pKa = 11.84SRR854 pKa = 11.84AEE856 pKa = 3.48EE857 pKa = 3.85RR858 pKa = 11.84ALYY861 pKa = 7.48STPRR865 pKa = 11.84SAVEE869 pKa = 3.89GLVLSLPVRR878 pKa = 11.84AKK880 pKa = 10.59RR881 pKa = 11.84LLAEE885 pKa = 4.09ALGWVV890 pKa = 3.71
Molecular weight: 99.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1349 |
459 |
890 |
674.5 |
75.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.192 ± 1.318 | 2.076 ± 0.247 |
4.448 ± 0.158 | 6.004 ± 0.543 |
3.484 ± 0.099 | 7.784 ± 0.527 |
2.52 ± 0.543 | 2.52 ± 0.057 |
3.632 ± 0.167 | 10.526 ± 0.631 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.038 ± 0.276 | 2.001 ± 0.081 |
8.08 ± 0.69 | 3.113 ± 0.87 |
12.305 ± 0.551 | 6.301 ± 0.008 |
4.448 ± 0.541 | 7.042 ± 0.332 |
2.224 ± 0.62 | 1.26 ± 0.178 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
