
Candidatus Annandia adelgestsuga
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Enterobacteriaceae incertae sedis; ant, tsetse, mealybug, aphid, etc. endosymbionts; Candidatus Annandia
Average proteome isoelectric point is 8.94
Get precalculated fractions of proteins
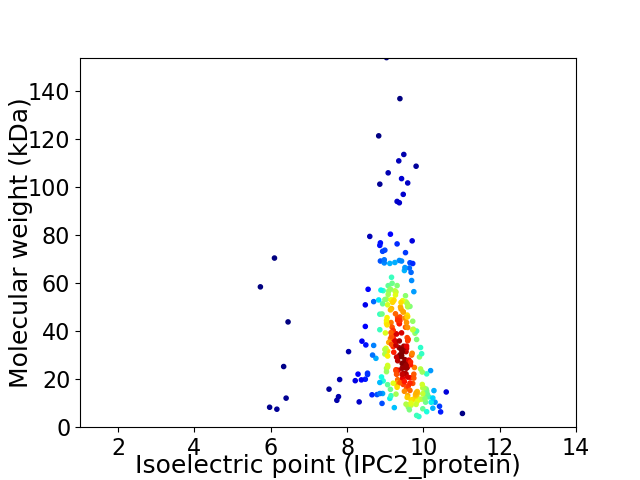
Virtual 2D-PAGE plot for 285 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9CLV2|A0A3Q9CLV2_9ENTR Methionine--tRNA ligase OS=Candidatus Annandia adelgestsuga OX=1302411 GN=metG PE=3 SV=1
MM1 pKa = 6.63TAKK4 pKa = 10.41KK5 pKa = 10.2IKK7 pKa = 9.93FGNDD11 pKa = 2.61ARR13 pKa = 11.84SKK15 pKa = 10.02ILKK18 pKa = 9.55GVNLLADD25 pKa = 3.83AVKK28 pKa = 9.58VTLGPKK34 pKa = 9.43GRR36 pKa = 11.84NAVIDD41 pKa = 3.92KK42 pKa = 11.07SFGAPIITKK51 pKa = 10.46DD52 pKa = 3.23GVTVARR58 pKa = 11.84EE59 pKa = 3.62IEE61 pKa = 4.11LDD63 pKa = 3.77DD64 pKa = 4.3KK65 pKa = 11.13FEE67 pKa = 4.52NMGAQMVKK75 pKa = 10.19EE76 pKa = 4.43VASKK80 pKa = 11.37ANDD83 pKa = 3.44AAGDD87 pKa = 3.92GTTTATLLAQSIVNEE102 pKa = 4.04GLKK105 pKa = 10.61AVAAGMNPMDD115 pKa = 5.27LKK117 pKa = 10.96RR118 pKa = 11.84GIDD121 pKa = 3.39KK122 pKa = 10.97AVILAVRR129 pKa = 11.84EE130 pKa = 3.99LKK132 pKa = 10.44IISVPCEE139 pKa = 3.71DD140 pKa = 4.41SKK142 pKa = 11.87SITQVGTISANADD155 pKa = 3.45EE156 pKa = 5.06NVGKK160 pKa = 10.5LISQAMSKK168 pKa = 10.08VGKK171 pKa = 9.73EE172 pKa = 3.81GVITVEE178 pKa = 4.33EE179 pKa = 4.62GTGLDD184 pKa = 3.95DD185 pKa = 4.36EE186 pKa = 5.38LDD188 pKa = 3.84VVEE191 pKa = 6.18GMQFDD196 pKa = 3.76RR197 pKa = 11.84GYY199 pKa = 10.76LSPYY203 pKa = 9.94FINKK207 pKa = 8.4QDD209 pKa = 3.55NGTVEE214 pKa = 4.42LDD216 pKa = 3.19NPYY219 pKa = 10.58ILLVEE224 pKa = 4.38KK225 pKa = 10.27KK226 pKa = 9.77ISNIRR231 pKa = 11.84EE232 pKa = 3.97LLSILEE238 pKa = 4.32NVAKK242 pKa = 10.55SNKK245 pKa = 8.83PMLIIAEE252 pKa = 4.68DD253 pKa = 3.63IEE255 pKa = 4.92GEE257 pKa = 4.08ALATLVVNNMRR268 pKa = 11.84GIVKK272 pKa = 9.77VAAVKK277 pKa = 10.73APGFGDD283 pKa = 3.21RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 11.2SMLQDD291 pKa = 3.1IAILTGGTVISEE303 pKa = 4.37EE304 pKa = 3.89IGIEE308 pKa = 3.87LEE310 pKa = 4.74KK311 pKa = 10.91INLEE315 pKa = 4.08NLGQAKK321 pKa = 9.64KK322 pKa = 11.06VIINKK327 pKa = 10.02DD328 pKa = 2.97NTTIIDD334 pKa = 4.67GIGKK338 pKa = 8.9EE339 pKa = 4.15KK340 pKa = 10.41KK341 pKa = 9.79IKK343 pKa = 10.59DD344 pKa = 3.48RR345 pKa = 11.84VKK347 pKa = 10.59QIHH350 pKa = 4.81QQIKK354 pKa = 9.5EE355 pKa = 4.06ATSDD359 pKa = 3.46YY360 pKa = 11.27DD361 pKa = 3.8KK362 pKa = 11.47EE363 pKa = 4.15KK364 pKa = 10.37LQEE367 pKa = 4.01RR368 pKa = 11.84VAKK371 pKa = 10.23LAGGVAVLKK380 pKa = 10.81VGAATEE386 pKa = 4.21VEE388 pKa = 4.37MKK390 pKa = 10.02EE391 pKa = 3.77KK392 pKa = 10.23KK393 pKa = 10.49ARR395 pKa = 11.84VEE397 pKa = 4.2DD398 pKa = 3.97ALHH401 pKa = 5.68ATRR404 pKa = 11.84AAVEE408 pKa = 4.18EE409 pKa = 4.64GVVAGGGVALIRR421 pKa = 11.84VASRR425 pKa = 11.84ISHH428 pKa = 6.46MIGQNEE434 pKa = 4.16DD435 pKa = 3.23QNVGIRR441 pKa = 11.84IAIRR445 pKa = 11.84AMEE448 pKa = 4.4APLRR452 pKa = 11.84QIIANAGEE460 pKa = 4.23EE461 pKa = 4.03PSVISNAIKK470 pKa = 10.41SGKK473 pKa = 9.57GNYY476 pKa = 9.11GYY478 pKa = 10.9NAATSEE484 pKa = 4.4YY485 pKa = 10.58GNMINFGILDD495 pKa = 3.74PTKK498 pKa = 8.82VTRR501 pKa = 11.84SALQYY506 pKa = 9.29ASSVAGLMITTEE518 pKa = 4.14CMITDD523 pKa = 3.6SSKK526 pKa = 11.2EE527 pKa = 4.05EE528 pKa = 4.1KK529 pKa = 10.51NDD531 pKa = 3.69TNNSIPSGGMGGMGGMMM548 pKa = 4.58
MM1 pKa = 6.63TAKK4 pKa = 10.41KK5 pKa = 10.2IKK7 pKa = 9.93FGNDD11 pKa = 2.61ARR13 pKa = 11.84SKK15 pKa = 10.02ILKK18 pKa = 9.55GVNLLADD25 pKa = 3.83AVKK28 pKa = 9.58VTLGPKK34 pKa = 9.43GRR36 pKa = 11.84NAVIDD41 pKa = 3.92KK42 pKa = 11.07SFGAPIITKK51 pKa = 10.46DD52 pKa = 3.23GVTVARR58 pKa = 11.84EE59 pKa = 3.62IEE61 pKa = 4.11LDD63 pKa = 3.77DD64 pKa = 4.3KK65 pKa = 11.13FEE67 pKa = 4.52NMGAQMVKK75 pKa = 10.19EE76 pKa = 4.43VASKK80 pKa = 11.37ANDD83 pKa = 3.44AAGDD87 pKa = 3.92GTTTATLLAQSIVNEE102 pKa = 4.04GLKK105 pKa = 10.61AVAAGMNPMDD115 pKa = 5.27LKK117 pKa = 10.96RR118 pKa = 11.84GIDD121 pKa = 3.39KK122 pKa = 10.97AVILAVRR129 pKa = 11.84EE130 pKa = 3.99LKK132 pKa = 10.44IISVPCEE139 pKa = 3.71DD140 pKa = 4.41SKK142 pKa = 11.87SITQVGTISANADD155 pKa = 3.45EE156 pKa = 5.06NVGKK160 pKa = 10.5LISQAMSKK168 pKa = 10.08VGKK171 pKa = 9.73EE172 pKa = 3.81GVITVEE178 pKa = 4.33EE179 pKa = 4.62GTGLDD184 pKa = 3.95DD185 pKa = 4.36EE186 pKa = 5.38LDD188 pKa = 3.84VVEE191 pKa = 6.18GMQFDD196 pKa = 3.76RR197 pKa = 11.84GYY199 pKa = 10.76LSPYY203 pKa = 9.94FINKK207 pKa = 8.4QDD209 pKa = 3.55NGTVEE214 pKa = 4.42LDD216 pKa = 3.19NPYY219 pKa = 10.58ILLVEE224 pKa = 4.38KK225 pKa = 10.27KK226 pKa = 9.77ISNIRR231 pKa = 11.84EE232 pKa = 3.97LLSILEE238 pKa = 4.32NVAKK242 pKa = 10.55SNKK245 pKa = 8.83PMLIIAEE252 pKa = 4.68DD253 pKa = 3.63IEE255 pKa = 4.92GEE257 pKa = 4.08ALATLVVNNMRR268 pKa = 11.84GIVKK272 pKa = 9.77VAAVKK277 pKa = 10.73APGFGDD283 pKa = 3.21RR284 pKa = 11.84RR285 pKa = 11.84KK286 pKa = 11.2SMLQDD291 pKa = 3.1IAILTGGTVISEE303 pKa = 4.37EE304 pKa = 3.89IGIEE308 pKa = 3.87LEE310 pKa = 4.74KK311 pKa = 10.91INLEE315 pKa = 4.08NLGQAKK321 pKa = 9.64KK322 pKa = 11.06VIINKK327 pKa = 10.02DD328 pKa = 2.97NTTIIDD334 pKa = 4.67GIGKK338 pKa = 8.9EE339 pKa = 4.15KK340 pKa = 10.41KK341 pKa = 9.79IKK343 pKa = 10.59DD344 pKa = 3.48RR345 pKa = 11.84VKK347 pKa = 10.59QIHH350 pKa = 4.81QQIKK354 pKa = 9.5EE355 pKa = 4.06ATSDD359 pKa = 3.46YY360 pKa = 11.27DD361 pKa = 3.8KK362 pKa = 11.47EE363 pKa = 4.15KK364 pKa = 10.37LQEE367 pKa = 4.01RR368 pKa = 11.84VAKK371 pKa = 10.23LAGGVAVLKK380 pKa = 10.81VGAATEE386 pKa = 4.21VEE388 pKa = 4.37MKK390 pKa = 10.02EE391 pKa = 3.77KK392 pKa = 10.23KK393 pKa = 10.49ARR395 pKa = 11.84VEE397 pKa = 4.2DD398 pKa = 3.97ALHH401 pKa = 5.68ATRR404 pKa = 11.84AAVEE408 pKa = 4.18EE409 pKa = 4.64GVVAGGGVALIRR421 pKa = 11.84VASRR425 pKa = 11.84ISHH428 pKa = 6.46MIGQNEE434 pKa = 4.16DD435 pKa = 3.23QNVGIRR441 pKa = 11.84IAIRR445 pKa = 11.84AMEE448 pKa = 4.4APLRR452 pKa = 11.84QIIANAGEE460 pKa = 4.23EE461 pKa = 4.03PSVISNAIKK470 pKa = 10.41SGKK473 pKa = 9.57GNYY476 pKa = 9.11GYY478 pKa = 10.9NAATSEE484 pKa = 4.4YY485 pKa = 10.58GNMINFGILDD495 pKa = 3.74PTKK498 pKa = 8.82VTRR501 pKa = 11.84SALQYY506 pKa = 9.29ASSVAGLMITTEE518 pKa = 4.14CMITDD523 pKa = 3.6SSKK526 pKa = 11.2EE527 pKa = 4.05EE528 pKa = 4.1KK529 pKa = 10.51NDD531 pKa = 3.69TNNSIPSGGMGGMGGMMM548 pKa = 4.58
Molecular weight: 58.42 kDa
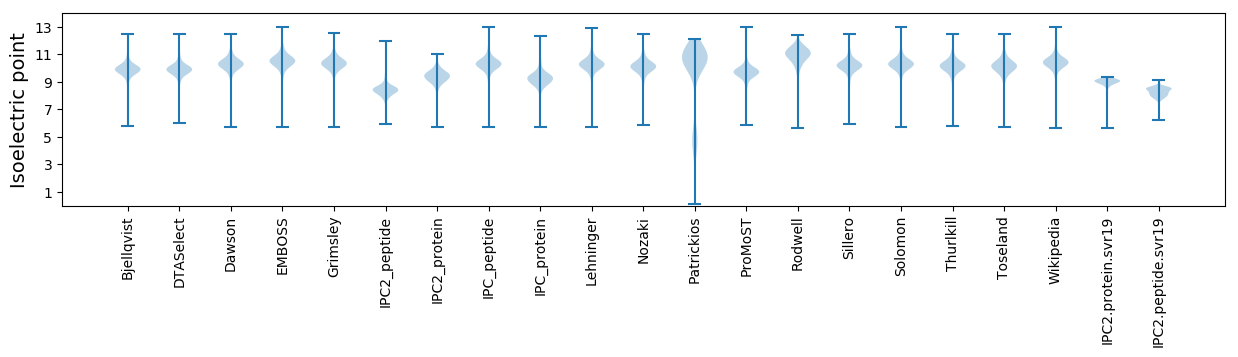
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S9J7J5|A0A3S9J7J5_9ENTR 3-isopropylmalate dehydratase large subunit OS=Candidatus Annandia adelgestsuga OX=1302411 GN=leuC PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.32KK12 pKa = 9.09KK13 pKa = 9.63RR14 pKa = 11.84SHH16 pKa = 5.84GFRR19 pKa = 11.84YY20 pKa = 10.12RR21 pKa = 11.84MSTKK25 pKa = 9.94NGRR28 pKa = 11.84NILSKK33 pKa = 10.55RR34 pKa = 11.84RR35 pKa = 11.84NKK37 pKa = 9.76KK38 pKa = 10.16RR39 pKa = 11.84LFLTISSKK47 pKa = 11.21
MM1 pKa = 7.45KK2 pKa = 9.58RR3 pKa = 11.84TFQPSLIKK11 pKa = 10.32KK12 pKa = 9.09KK13 pKa = 9.63RR14 pKa = 11.84SHH16 pKa = 5.84GFRR19 pKa = 11.84YY20 pKa = 10.12RR21 pKa = 11.84MSTKK25 pKa = 9.94NGRR28 pKa = 11.84NILSKK33 pKa = 10.55RR34 pKa = 11.84RR35 pKa = 11.84NKK37 pKa = 9.76KK38 pKa = 10.16RR39 pKa = 11.84LFLTISSKK47 pKa = 11.21
Molecular weight: 5.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
90704 |
38 |
1336 |
318.3 |
36.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.728 ± 0.114 | 1.274 ± 0.048 |
3.291 ± 0.079 | 3.755 ± 0.108 |
5.439 ± 0.147 | 5.025 ± 0.134 |
1.428 ± 0.045 | 15.149 ± 0.194 |
14.082 ± 0.251 | 9.431 ± 0.136 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.011 ± 0.052 | 10.228 ± 0.187 |
2.581 ± 0.068 | 1.843 ± 0.051 |
2.687 ± 0.081 | 6.34 ± 0.109 |
3.57 ± 0.08 | 3.361 ± 0.099 |
0.766 ± 0.043 | 5.01 ± 0.108 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
