
Desulfosporosinus fructosivorans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Peptococcaceae; Desulfosporosinus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
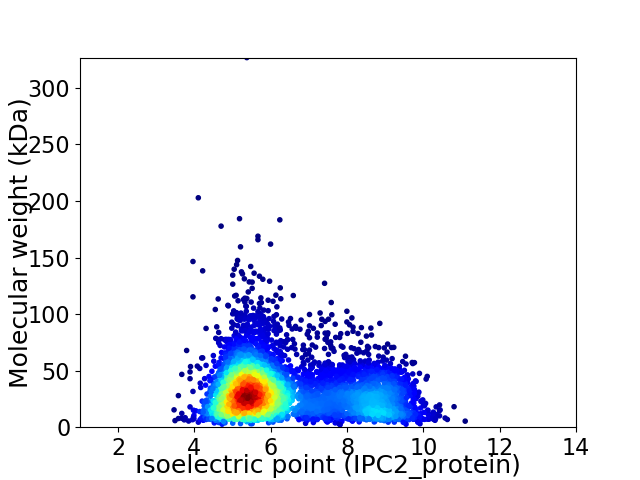
Virtual 2D-PAGE plot for 5570 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z0QZI7|A0A4Z0QZI7_9FIRM Ferredoxin family protein OS=Desulfosporosinus fructosivorans OX=2018669 GN=E4K67_25505 PE=4 SV=1
MM1 pKa = 6.88MRR3 pKa = 11.84ALFAGVSGLQAHH15 pKa = 5.45QKK17 pKa = 10.4KK18 pKa = 9.42MDD20 pKa = 3.81VIGNNIANVNTVAYY34 pKa = 8.5KK35 pKa = 10.33ASRR38 pKa = 11.84VTFSDD43 pKa = 3.62VLSQTLSKK51 pKa = 10.06ATAANSDD58 pKa = 3.88SGVGGTNPKK67 pKa = 10.34QIGLGVGVASIDD79 pKa = 3.43MLMTDD84 pKa = 4.67GGTEE88 pKa = 4.12STGNTTDD95 pKa = 4.58LSLEE99 pKa = 4.01GDD101 pKa = 3.66GFFIVRR107 pKa = 11.84NGVTGSYY114 pKa = 9.1MFTRR118 pKa = 11.84AGDD121 pKa = 3.53FTVDD125 pKa = 3.12EE126 pKa = 4.96KK127 pKa = 11.91GNLVTSDD134 pKa = 2.91GMNVYY139 pKa = 9.44GWSSYY144 pKa = 7.4STNAEE149 pKa = 3.27GDD151 pKa = 3.91YY152 pKa = 11.4EE153 pKa = 4.84FDD155 pKa = 3.2TDD157 pKa = 4.86NEE159 pKa = 4.18VEE161 pKa = 4.77AINIYY166 pKa = 10.97ADD168 pKa = 3.62AYY170 pKa = 10.66NGNKK174 pKa = 9.89KK175 pKa = 9.63ILSAQATEE183 pKa = 4.04NVVFSGSLDD192 pKa = 3.5SSEE195 pKa = 4.36EE196 pKa = 4.13AIEE199 pKa = 6.04DD200 pKa = 3.48IDD202 pKa = 6.34DD203 pKa = 4.09DD204 pKa = 4.49TEE206 pKa = 4.14PQYY209 pKa = 8.29TTSMTVYY216 pKa = 10.31DD217 pKa = 3.95SLGNEE222 pKa = 4.25HH223 pKa = 7.27EE224 pKa = 4.34LTVNYY229 pKa = 9.76IKK231 pKa = 10.73SDD233 pKa = 3.45TNTWDD238 pKa = 3.36CYY240 pKa = 7.42VTYY243 pKa = 10.66DD244 pKa = 4.31SGEE247 pKa = 4.04TDD249 pKa = 3.33TEE251 pKa = 3.94GDD253 pKa = 4.0AILTRR258 pKa = 11.84VDD260 pKa = 2.92IGEE263 pKa = 4.27LQFDD267 pKa = 4.06EE268 pKa = 4.92YY269 pKa = 11.54GDD271 pKa = 3.65IVEE274 pKa = 5.91DD275 pKa = 3.67DD276 pKa = 3.35TSYY279 pKa = 8.59PTTQVLTITPTTGATDD295 pKa = 4.08LEE297 pKa = 4.34ITLDD301 pKa = 3.81FTNLSTGADD310 pKa = 3.76DD311 pKa = 4.38SSVEE315 pKa = 3.85AGEE318 pKa = 3.79IDD320 pKa = 4.44GYY322 pKa = 11.51SSGTLEE328 pKa = 5.91DD329 pKa = 3.97ISIDD333 pKa = 3.61SNGIIMGVYY342 pKa = 10.42SNGSQQPLGMIGIAQFANSSGLQKK366 pKa = 9.77TGSGYY371 pKa = 11.2YY372 pKa = 10.08SATANSGQFTNGVSANGAISSGTLEE397 pKa = 4.2MSNVDD402 pKa = 3.48LSYY405 pKa = 10.98EE406 pKa = 4.48FSQMITTQRR415 pKa = 11.84GFQANSTLITTADD428 pKa = 3.47EE429 pKa = 4.1MLEE432 pKa = 3.93TLINMKK438 pKa = 10.29RR439 pKa = 3.25
MM1 pKa = 6.88MRR3 pKa = 11.84ALFAGVSGLQAHH15 pKa = 5.45QKK17 pKa = 10.4KK18 pKa = 9.42MDD20 pKa = 3.81VIGNNIANVNTVAYY34 pKa = 8.5KK35 pKa = 10.33ASRR38 pKa = 11.84VTFSDD43 pKa = 3.62VLSQTLSKK51 pKa = 10.06ATAANSDD58 pKa = 3.88SGVGGTNPKK67 pKa = 10.34QIGLGVGVASIDD79 pKa = 3.43MLMTDD84 pKa = 4.67GGTEE88 pKa = 4.12STGNTTDD95 pKa = 4.58LSLEE99 pKa = 4.01GDD101 pKa = 3.66GFFIVRR107 pKa = 11.84NGVTGSYY114 pKa = 9.1MFTRR118 pKa = 11.84AGDD121 pKa = 3.53FTVDD125 pKa = 3.12EE126 pKa = 4.96KK127 pKa = 11.91GNLVTSDD134 pKa = 2.91GMNVYY139 pKa = 9.44GWSSYY144 pKa = 7.4STNAEE149 pKa = 3.27GDD151 pKa = 3.91YY152 pKa = 11.4EE153 pKa = 4.84FDD155 pKa = 3.2TDD157 pKa = 4.86NEE159 pKa = 4.18VEE161 pKa = 4.77AINIYY166 pKa = 10.97ADD168 pKa = 3.62AYY170 pKa = 10.66NGNKK174 pKa = 9.89KK175 pKa = 9.63ILSAQATEE183 pKa = 4.04NVVFSGSLDD192 pKa = 3.5SSEE195 pKa = 4.36EE196 pKa = 4.13AIEE199 pKa = 6.04DD200 pKa = 3.48IDD202 pKa = 6.34DD203 pKa = 4.09DD204 pKa = 4.49TEE206 pKa = 4.14PQYY209 pKa = 8.29TTSMTVYY216 pKa = 10.31DD217 pKa = 3.95SLGNEE222 pKa = 4.25HH223 pKa = 7.27EE224 pKa = 4.34LTVNYY229 pKa = 9.76IKK231 pKa = 10.73SDD233 pKa = 3.45TNTWDD238 pKa = 3.36CYY240 pKa = 7.42VTYY243 pKa = 10.66DD244 pKa = 4.31SGEE247 pKa = 4.04TDD249 pKa = 3.33TEE251 pKa = 3.94GDD253 pKa = 4.0AILTRR258 pKa = 11.84VDD260 pKa = 2.92IGEE263 pKa = 4.27LQFDD267 pKa = 4.06EE268 pKa = 4.92YY269 pKa = 11.54GDD271 pKa = 3.65IVEE274 pKa = 5.91DD275 pKa = 3.67DD276 pKa = 3.35TSYY279 pKa = 8.59PTTQVLTITPTTGATDD295 pKa = 4.08LEE297 pKa = 4.34ITLDD301 pKa = 3.81FTNLSTGADD310 pKa = 3.76DD311 pKa = 4.38SSVEE315 pKa = 3.85AGEE318 pKa = 3.79IDD320 pKa = 4.44GYY322 pKa = 11.51SSGTLEE328 pKa = 5.91DD329 pKa = 3.97ISIDD333 pKa = 3.61SNGIIMGVYY342 pKa = 10.42SNGSQQPLGMIGIAQFANSSGLQKK366 pKa = 9.77TGSGYY371 pKa = 11.2YY372 pKa = 10.08SATANSGQFTNGVSANGAISSGTLEE397 pKa = 4.2MSNVDD402 pKa = 3.48LSYY405 pKa = 10.98EE406 pKa = 4.48FSQMITTQRR415 pKa = 11.84GFQANSTLITTADD428 pKa = 3.47EE429 pKa = 4.1MLEE432 pKa = 3.93TLINMKK438 pKa = 10.29RR439 pKa = 3.25
Molecular weight: 46.69 kDa
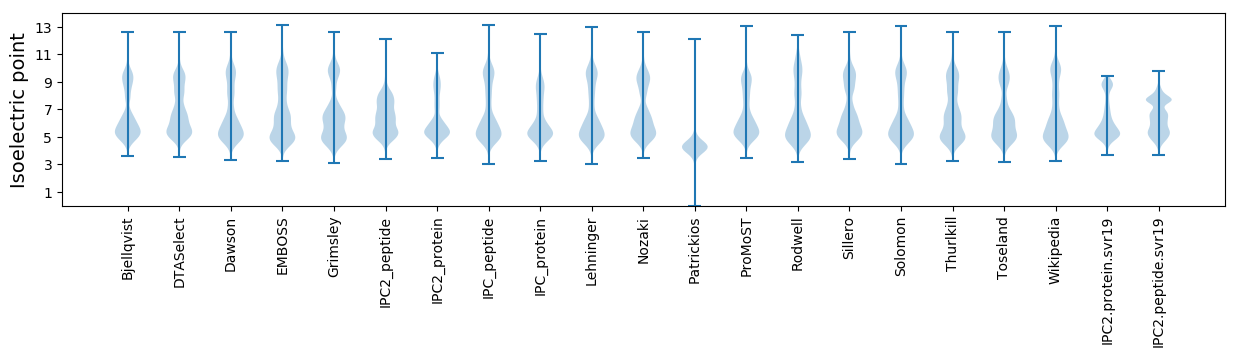
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z0R4W4|A0A4Z0R4W4_9FIRM Protein-glutamate O-methyltransferase OS=Desulfosporosinus fructosivorans OX=2018669 GN=E4K67_18385 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.19QPKK8 pKa = 8.78NRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.44KK13 pKa = 10.09RR14 pKa = 11.84VHH16 pKa = 5.93GFLSRR21 pKa = 11.84MSTPTGRR28 pKa = 11.84NVIKK32 pKa = 10.5RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 8.8KK41 pKa = 10.81LSVV44 pKa = 3.15
MM1 pKa = 7.36KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 9.19QPKK8 pKa = 8.78NRR10 pKa = 11.84RR11 pKa = 11.84HH12 pKa = 5.44KK13 pKa = 10.09RR14 pKa = 11.84VHH16 pKa = 5.93GFLSRR21 pKa = 11.84MSTPTGRR28 pKa = 11.84NVIKK32 pKa = 10.5RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 8.8KK41 pKa = 10.81LSVV44 pKa = 3.15
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1667623 |
22 |
2862 |
299.4 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.669 ± 0.041 | 1.166 ± 0.015 |
4.924 ± 0.027 | 6.75 ± 0.033 |
4.069 ± 0.025 | 7.452 ± 0.032 |
1.809 ± 0.014 | 7.643 ± 0.026 |
6.109 ± 0.031 | 10.371 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.763 ± 0.017 | 4.123 ± 0.024 |
3.862 ± 0.019 | 3.669 ± 0.019 |
4.502 ± 0.024 | 6.128 ± 0.029 |
5.426 ± 0.028 | 7.298 ± 0.028 |
1.047 ± 0.013 | 3.221 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
