
Serpentinomonas raichei
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Serpentinomonas
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
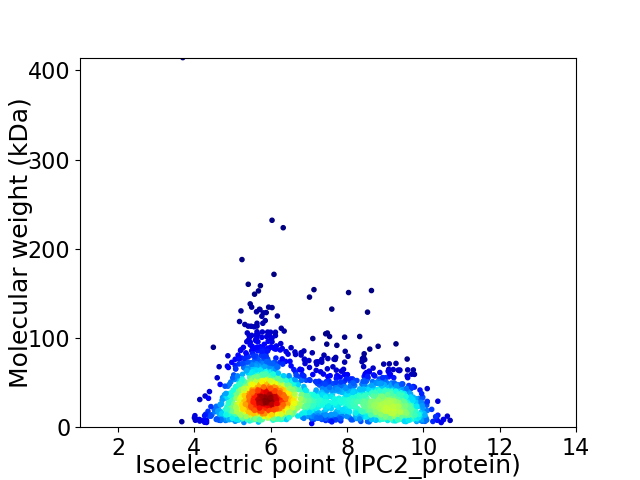
Virtual 2D-PAGE plot for 2363 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
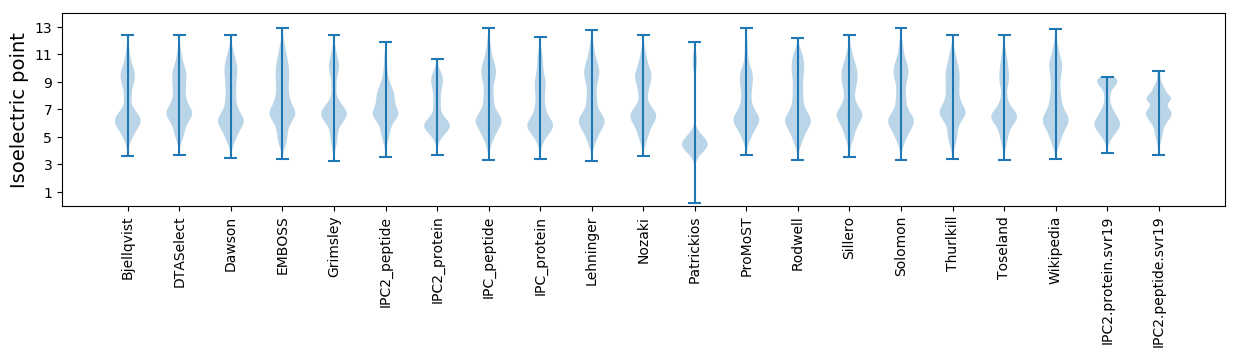
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A060NT44|A0A060NT44_9BURK Thioredoxin domain-containing protein OS=Serpentinomonas raichei OX=1458425 GN=SRAA_2225 PE=3 SV=1
MM1 pKa = 7.64AATTLIMKK9 pKa = 7.59TFTAIVEE16 pKa = 4.47KK17 pKa = 10.17CTEE20 pKa = 3.73TGLYY24 pKa = 9.0VGYY27 pKa = 10.94VPGFPGAHH35 pKa = 5.05TQGTSLDD42 pKa = 3.82EE43 pKa = 4.23LQQNLQEE50 pKa = 4.33VVSMLLEE57 pKa = 4.91DD58 pKa = 4.57GEE60 pKa = 4.75PVLDD64 pKa = 3.77AQFVGTQQLAIAA76 pKa = 4.89
MM1 pKa = 7.64AATTLIMKK9 pKa = 7.59TFTAIVEE16 pKa = 4.47KK17 pKa = 10.17CTEE20 pKa = 3.73TGLYY24 pKa = 9.0VGYY27 pKa = 10.94VPGFPGAHH35 pKa = 5.05TQGTSLDD42 pKa = 3.82EE43 pKa = 4.23LQQNLQEE50 pKa = 4.33VVSMLLEE57 pKa = 4.91DD58 pKa = 4.57GEE60 pKa = 4.75PVLDD64 pKa = 3.77AQFVGTQQLAIAA76 pKa = 4.89
Molecular weight: 8.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A060NF26|A0A060NF26_9BURK Beta sliding clamp OS=Serpentinomonas raichei OX=1458425 GN=SRAA_0002 PE=3 SV=1
MM1 pKa = 7.46EE2 pKa = 5.58PALNHH7 pKa = 6.89ANLTLSGRR15 pKa = 11.84EE16 pKa = 3.93LARR19 pKa = 11.84MRR21 pKa = 11.84RR22 pKa = 11.84LAMATEE28 pKa = 4.51GKK30 pKa = 10.19VGAANRR36 pKa = 11.84VAAAVRR42 pKa = 11.84ASQAAAQAVQAAARR56 pKa = 11.84RR57 pKa = 11.84DD58 pKa = 3.66EE59 pKa = 4.35AQCGCSGAADD69 pKa = 4.13AACQCDD75 pKa = 3.52VAAAVSAPASQTKK88 pKa = 9.35AACALPNGRR97 pKa = 11.84TLARR101 pKa = 11.84ARR103 pKa = 11.84RR104 pKa = 11.84QTLAQEE110 pKa = 4.53GKK112 pKa = 10.69VGIQKK117 pKa = 9.54VANASRR123 pKa = 11.84IAATMPDD130 pKa = 3.24RR131 pKa = 11.84ADD133 pKa = 3.07WQTALVQGVTGRR145 pKa = 11.84QLAMQKK151 pKa = 10.4RR152 pKa = 11.84IVQSLAGRR160 pKa = 11.84TEE162 pKa = 4.1AQHH165 pKa = 6.45SSGNRR170 pKa = 11.84SVARR174 pKa = 11.84SRR176 pKa = 11.84SRR178 pKa = 11.84PVAEE182 pKa = 3.88RR183 pKa = 11.84TEE185 pKa = 4.21VGHH188 pKa = 5.62TLSGQEE194 pKa = 3.56VSGIQVEE201 pKa = 4.47RR202 pKa = 11.84SSKK205 pKa = 9.22VTGQEE210 pKa = 3.5PGTCRR215 pKa = 11.84NVTGTEE221 pKa = 4.18YY222 pKa = 10.18IGMEE226 pKa = 4.48QYY228 pKa = 10.41QAWCEE233 pKa = 4.31SKK235 pKa = 10.34PQPRR239 pKa = 11.84PPKK242 pKa = 10.14VGLSQTTGQQQSISGTEE259 pKa = 3.77VDD261 pKa = 3.69PQPRR265 pKa = 11.84VTGNQTGVCLGITGTQYY282 pKa = 11.47LSDD285 pKa = 3.79LTTRR289 pKa = 11.84LCQEE293 pKa = 4.0QPLQGPHH300 pKa = 6.33KK301 pKa = 9.57VSVMSSRR308 pKa = 11.84GQQTVTGVVVNGGSKK323 pKa = 8.51VTGNEE328 pKa = 3.52AGMQRR333 pKa = 11.84PITGTQYY340 pKa = 11.52ARR342 pKa = 11.84ALQAAPSRR350 pKa = 11.84MPASEE355 pKa = 4.21ARR357 pKa = 11.84HH358 pKa = 5.71ALPEE362 pKa = 4.01PVQVQRR368 pKa = 11.84QSASWRR374 pKa = 11.84TQSLTGDD381 pKa = 3.43RR382 pKa = 11.84PGIGGGGVTGDD393 pKa = 3.42EE394 pKa = 4.48RR395 pKa = 11.84GACEE399 pKa = 5.31PITGTPFIGPDD410 pKa = 3.27NQYY413 pKa = 10.9AACTIDD419 pKa = 3.95SAWLTRR425 pKa = 11.84HH426 pKa = 6.5PEE428 pKa = 4.07LAVEE432 pKa = 4.72TPPAAPKK439 pKa = 10.2GFSIARR445 pKa = 11.84PQRR448 pKa = 11.84GQPAGRR454 pKa = 11.84ATTPQSAAPRR464 pKa = 11.84NQVTGVAYY472 pKa = 10.33GNSEE476 pKa = 4.9RR477 pKa = 11.84ITGPGFKK484 pKa = 10.19AQGVITGTPEE494 pKa = 3.56FRR496 pKa = 11.84HH497 pKa = 6.23GGQSLDD503 pKa = 3.45SRR505 pKa = 11.84RR506 pKa = 11.84SSARR510 pKa = 11.84PATPASPPAVQPPASEE526 pKa = 4.1RR527 pKa = 11.84LSGEE531 pKa = 3.59GRR533 pKa = 11.84QQGSRR538 pKa = 11.84LTGDD542 pKa = 2.8SWSSSRR548 pKa = 11.84HH549 pKa = 3.99ISGTDD554 pKa = 3.34GVSAQSRR561 pKa = 11.84NPTQRR566 pKa = 11.84GQPRR570 pKa = 11.84AMGMGMDD577 pKa = 3.45ARR579 pKa = 11.84SVRR582 pKa = 11.84EE583 pKa = 3.69QARR586 pKa = 11.84AEE588 pKa = 4.24VPPSPVTGSSGNTGRR603 pKa = 11.84GAMVTISGGARR614 pKa = 11.84GG615 pKa = 3.54
MM1 pKa = 7.46EE2 pKa = 5.58PALNHH7 pKa = 6.89ANLTLSGRR15 pKa = 11.84EE16 pKa = 3.93LARR19 pKa = 11.84MRR21 pKa = 11.84RR22 pKa = 11.84LAMATEE28 pKa = 4.51GKK30 pKa = 10.19VGAANRR36 pKa = 11.84VAAAVRR42 pKa = 11.84ASQAAAQAVQAAARR56 pKa = 11.84RR57 pKa = 11.84DD58 pKa = 3.66EE59 pKa = 4.35AQCGCSGAADD69 pKa = 4.13AACQCDD75 pKa = 3.52VAAAVSAPASQTKK88 pKa = 9.35AACALPNGRR97 pKa = 11.84TLARR101 pKa = 11.84ARR103 pKa = 11.84RR104 pKa = 11.84QTLAQEE110 pKa = 4.53GKK112 pKa = 10.69VGIQKK117 pKa = 9.54VANASRR123 pKa = 11.84IAATMPDD130 pKa = 3.24RR131 pKa = 11.84ADD133 pKa = 3.07WQTALVQGVTGRR145 pKa = 11.84QLAMQKK151 pKa = 10.4RR152 pKa = 11.84IVQSLAGRR160 pKa = 11.84TEE162 pKa = 4.1AQHH165 pKa = 6.45SSGNRR170 pKa = 11.84SVARR174 pKa = 11.84SRR176 pKa = 11.84SRR178 pKa = 11.84PVAEE182 pKa = 3.88RR183 pKa = 11.84TEE185 pKa = 4.21VGHH188 pKa = 5.62TLSGQEE194 pKa = 3.56VSGIQVEE201 pKa = 4.47RR202 pKa = 11.84SSKK205 pKa = 9.22VTGQEE210 pKa = 3.5PGTCRR215 pKa = 11.84NVTGTEE221 pKa = 4.18YY222 pKa = 10.18IGMEE226 pKa = 4.48QYY228 pKa = 10.41QAWCEE233 pKa = 4.31SKK235 pKa = 10.34PQPRR239 pKa = 11.84PPKK242 pKa = 10.14VGLSQTTGQQQSISGTEE259 pKa = 3.77VDD261 pKa = 3.69PQPRR265 pKa = 11.84VTGNQTGVCLGITGTQYY282 pKa = 11.47LSDD285 pKa = 3.79LTTRR289 pKa = 11.84LCQEE293 pKa = 4.0QPLQGPHH300 pKa = 6.33KK301 pKa = 9.57VSVMSSRR308 pKa = 11.84GQQTVTGVVVNGGSKK323 pKa = 8.51VTGNEE328 pKa = 3.52AGMQRR333 pKa = 11.84PITGTQYY340 pKa = 11.52ARR342 pKa = 11.84ALQAAPSRR350 pKa = 11.84MPASEE355 pKa = 4.21ARR357 pKa = 11.84HH358 pKa = 5.71ALPEE362 pKa = 4.01PVQVQRR368 pKa = 11.84QSASWRR374 pKa = 11.84TQSLTGDD381 pKa = 3.43RR382 pKa = 11.84PGIGGGGVTGDD393 pKa = 3.42EE394 pKa = 4.48RR395 pKa = 11.84GACEE399 pKa = 5.31PITGTPFIGPDD410 pKa = 3.27NQYY413 pKa = 10.9AACTIDD419 pKa = 3.95SAWLTRR425 pKa = 11.84HH426 pKa = 6.5PEE428 pKa = 4.07LAVEE432 pKa = 4.72TPPAAPKK439 pKa = 10.2GFSIARR445 pKa = 11.84PQRR448 pKa = 11.84GQPAGRR454 pKa = 11.84ATTPQSAAPRR464 pKa = 11.84NQVTGVAYY472 pKa = 10.33GNSEE476 pKa = 4.9RR477 pKa = 11.84ITGPGFKK484 pKa = 10.19AQGVITGTPEE494 pKa = 3.56FRR496 pKa = 11.84HH497 pKa = 6.23GGQSLDD503 pKa = 3.45SRR505 pKa = 11.84RR506 pKa = 11.84SSARR510 pKa = 11.84PATPASPPAVQPPASEE526 pKa = 4.1RR527 pKa = 11.84LSGEE531 pKa = 3.59GRR533 pKa = 11.84QQGSRR538 pKa = 11.84LTGDD542 pKa = 2.8SWSSSRR548 pKa = 11.84HH549 pKa = 3.99ISGTDD554 pKa = 3.34GVSAQSRR561 pKa = 11.84NPTQRR566 pKa = 11.84GQPRR570 pKa = 11.84AMGMGMDD577 pKa = 3.45ARR579 pKa = 11.84SVRR582 pKa = 11.84EE583 pKa = 3.69QARR586 pKa = 11.84AEE588 pKa = 4.24VPPSPVTGSSGNTGRR603 pKa = 11.84GAMVTISGGARR614 pKa = 11.84GG615 pKa = 3.54
Molecular weight: 64.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
785481 |
41 |
4077 |
332.4 |
36.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.42 ± 0.076 | 1.016 ± 0.019 |
4.712 ± 0.041 | 5.363 ± 0.042 |
3.264 ± 0.028 | 8.054 ± 0.045 |
2.563 ± 0.031 | 3.997 ± 0.042 |
2.527 ± 0.049 | 12.183 ± 0.081 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.127 ± 0.025 | 2.33 ± 0.035 |
5.715 ± 0.04 | 5.255 ± 0.049 |
7.26 ± 0.048 | 5.025 ± 0.034 |
4.573 ± 0.044 | 7.049 ± 0.047 |
1.586 ± 0.025 | 1.98 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
