
Hubei virga-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
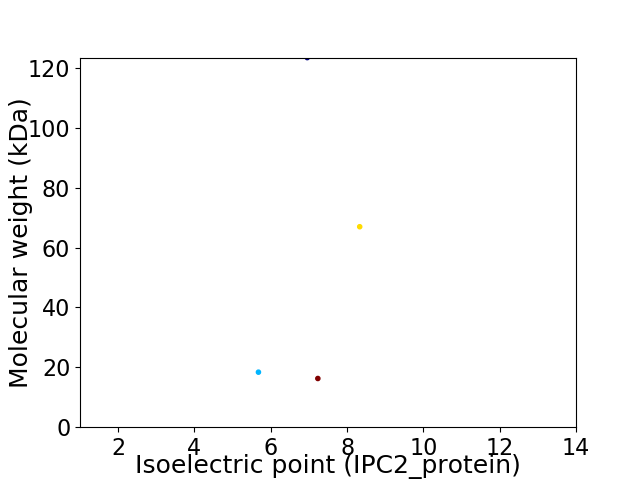
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJP3|A0A1L3KJP3_9VIRU Uncharacterized protein OS=Hubei virga-like virus 11 OX=1923326 PE=4 SV=1
MM1 pKa = 7.98PYY3 pKa = 9.07TWTTEE8 pKa = 3.71ALLLTTHH15 pKa = 6.38HH16 pKa = 5.77WVEE19 pKa = 4.14YY20 pKa = 9.25AAFMRR25 pKa = 11.84IIGSFKK31 pKa = 10.18FKK33 pKa = 10.48QFSVKK38 pKa = 10.13AVRR41 pKa = 11.84DD42 pKa = 3.82SMLTTLANLEE52 pKa = 4.06YY53 pKa = 8.91DD54 pKa = 3.59TPFNVTTRR62 pKa = 11.84FPEE65 pKa = 4.02DD66 pKa = 3.66KK67 pKa = 10.42IYY69 pKa = 10.54IDD71 pKa = 4.55LSHH74 pKa = 6.66SVPLKK79 pKa = 10.43ILQQLTKK86 pKa = 10.75ALDD89 pKa = 3.71FSDD92 pKa = 3.41RR93 pKa = 11.84QTEE96 pKa = 4.13KK97 pKa = 11.15GRR99 pKa = 11.84VTTDD103 pKa = 2.92QEE105 pKa = 4.29KK106 pKa = 10.79NRR108 pKa = 11.84THH110 pKa = 7.43SNFEE114 pKa = 3.98DD115 pKa = 3.49AKK117 pKa = 9.43VAFYY121 pKa = 9.62TAVDD125 pKa = 3.64QLIDD129 pKa = 4.59LIGPFPIEE137 pKa = 3.85AAHH140 pKa = 6.27VYY142 pKa = 10.03GVYY145 pKa = 9.96TRR147 pKa = 11.84TSFEE151 pKa = 3.96IEE153 pKa = 4.36KK154 pKa = 10.62GLVWAA159 pKa = 5.47
MM1 pKa = 7.98PYY3 pKa = 9.07TWTTEE8 pKa = 3.71ALLLTTHH15 pKa = 6.38HH16 pKa = 5.77WVEE19 pKa = 4.14YY20 pKa = 9.25AAFMRR25 pKa = 11.84IIGSFKK31 pKa = 10.18FKK33 pKa = 10.48QFSVKK38 pKa = 10.13AVRR41 pKa = 11.84DD42 pKa = 3.82SMLTTLANLEE52 pKa = 4.06YY53 pKa = 8.91DD54 pKa = 3.59TPFNVTTRR62 pKa = 11.84FPEE65 pKa = 4.02DD66 pKa = 3.66KK67 pKa = 10.42IYY69 pKa = 10.54IDD71 pKa = 4.55LSHH74 pKa = 6.66SVPLKK79 pKa = 10.43ILQQLTKK86 pKa = 10.75ALDD89 pKa = 3.71FSDD92 pKa = 3.41RR93 pKa = 11.84QTEE96 pKa = 4.13KK97 pKa = 11.15GRR99 pKa = 11.84VTTDD103 pKa = 2.92QEE105 pKa = 4.29KK106 pKa = 10.79NRR108 pKa = 11.84THH110 pKa = 7.43SNFEE114 pKa = 3.98DD115 pKa = 3.49AKK117 pKa = 9.43VAFYY121 pKa = 9.62TAVDD125 pKa = 3.64QLIDD129 pKa = 4.59LIGPFPIEE137 pKa = 3.85AAHH140 pKa = 6.27VYY142 pKa = 10.03GVYY145 pKa = 9.96TRR147 pKa = 11.84TSFEE151 pKa = 3.96IEE153 pKa = 4.36KK154 pKa = 10.62GLVWAA159 pKa = 5.47
Molecular weight: 18.39 kDa
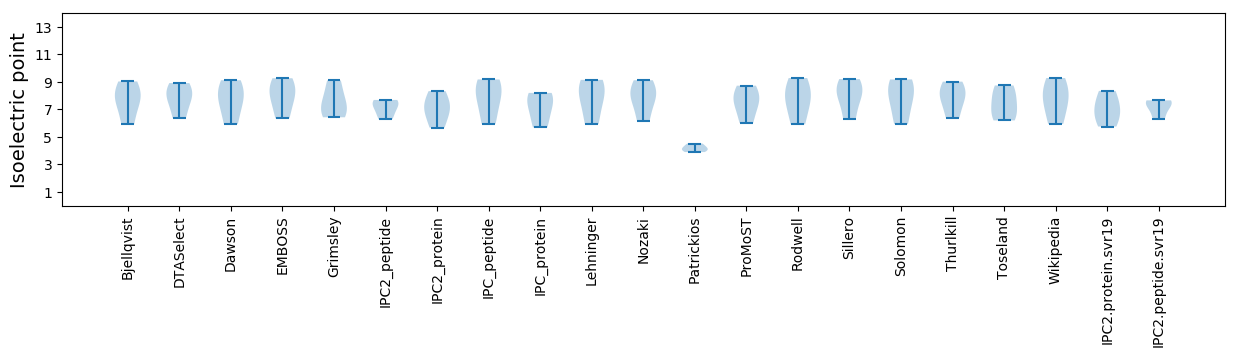
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJW5|A0A1L3KJW5_9VIRU Uncharacterized protein OS=Hubei virga-like virus 11 OX=1923326 PE=4 SV=1
MM1 pKa = 7.43SLVKK5 pKa = 10.64LQFANILLPLEE16 pKa = 4.49VSTSEE21 pKa = 3.55MDD23 pKa = 3.35QFKK26 pKa = 11.44LEE28 pKa = 4.8LINLLMTTFGSVNLFSSFTIFAVLSNNMKK57 pKa = 10.25LHH59 pKa = 6.75PLQLSIVDD67 pKa = 3.61HH68 pKa = 6.2TEE70 pKa = 3.91PGSIPMNRR78 pKa = 11.84EE79 pKa = 3.26VAEE82 pKa = 5.01FIAKK86 pKa = 8.32TFSDD90 pKa = 4.82RR91 pKa = 11.84ICRR94 pKa = 11.84TIGYY98 pKa = 9.34NIYY101 pKa = 8.93TLEE104 pKa = 4.23LAHH107 pKa = 7.15APINNQVDD115 pKa = 3.71DD116 pKa = 4.94DD117 pKa = 4.24FLSRR121 pKa = 11.84LSKK124 pKa = 11.06LNLSSSSSSDD134 pKa = 3.06SGNEE138 pKa = 3.91SFTLTIRR145 pKa = 11.84KK146 pKa = 8.77DD147 pKa = 3.31ASIGQIFDD155 pKa = 4.46SIINNAKK162 pKa = 10.34DD163 pKa = 4.02YY164 pKa = 11.55LPMKK168 pKa = 8.85TIKK171 pKa = 10.36DD172 pKa = 3.54ALKK175 pKa = 10.63ASVDD179 pKa = 3.56KK180 pKa = 10.03FTSPSNASATTTNGSRR196 pKa = 11.84ISRR199 pKa = 11.84SKK201 pKa = 11.18SDD203 pKa = 4.46DD204 pKa = 3.19SLKK207 pKa = 10.19RR208 pKa = 11.84DD209 pKa = 3.61KK210 pKa = 11.17EE211 pKa = 4.21STEE214 pKa = 3.61TDD216 pKa = 2.76KK217 pKa = 11.59RR218 pKa = 11.84MLGNMTASDD227 pKa = 4.65LLNAMSRR234 pKa = 11.84GCSNTCNITCPKK246 pKa = 10.07CEE248 pKa = 4.07YY249 pKa = 10.36IVVKK253 pKa = 10.96NNSTSNNGSITPARR267 pKa = 11.84KK268 pKa = 9.6SNNVSDD274 pKa = 4.26CLFYY278 pKa = 10.69TPQRR282 pKa = 11.84RR283 pKa = 11.84LTDD286 pKa = 3.54YY287 pKa = 7.45EE288 pKa = 4.42TKK290 pKa = 9.84FTVNYY295 pKa = 9.72NSQVYY300 pKa = 8.66NVDD303 pKa = 3.15MVYY306 pKa = 10.24MGMSEE311 pKa = 4.23RR312 pKa = 11.84NKK314 pKa = 9.94TIAFLDD320 pKa = 3.65VLLSLNNRR328 pKa = 11.84RR329 pKa = 11.84LFTNATEE336 pKa = 3.81NSSYY340 pKa = 10.81IKK342 pKa = 10.08RR343 pKa = 11.84HH344 pKa = 4.72YY345 pKa = 9.38KK346 pKa = 10.3RR347 pKa = 11.84NVPVLSTKK355 pKa = 10.42SSNATITTRR364 pKa = 11.84DD365 pKa = 3.67PNSIAMLRR373 pKa = 11.84RR374 pKa = 11.84VTIPPPSTPRR384 pKa = 11.84PLINTFVDD392 pKa = 3.44SFDD395 pKa = 3.49KK396 pKa = 11.68VEE398 pKa = 4.2INPRR402 pKa = 11.84THH404 pKa = 6.96SNIIPYY410 pKa = 9.86PSVNTGVINVRR421 pKa = 11.84NDD423 pKa = 3.49ALKK426 pKa = 10.5KK427 pKa = 10.29IYY429 pKa = 10.07DD430 pKa = 3.86SNGTLIMDD438 pKa = 4.21FTNITLEE445 pKa = 4.28IIRR448 pKa = 11.84EE449 pKa = 3.91GHH451 pKa = 5.06TYY453 pKa = 11.08KK454 pKa = 9.98FTTAKK459 pKa = 10.39GSYY462 pKa = 7.91NTAGEE467 pKa = 4.05FMKK470 pKa = 10.99YY471 pKa = 9.88IVDD474 pKa = 4.09AYY476 pKa = 10.45GLRR479 pKa = 11.84QTLEE483 pKa = 3.74LFMDD487 pKa = 4.62KK488 pKa = 10.52FIDD491 pKa = 3.83MPDD494 pKa = 3.24TDD496 pKa = 4.96FQALLDD502 pKa = 4.63GIIKK506 pKa = 8.89RR507 pKa = 11.84TRR509 pKa = 11.84LMKK512 pKa = 10.66RR513 pKa = 11.84MMHH516 pKa = 6.02NDD518 pKa = 3.04IYY520 pKa = 10.94RR521 pKa = 11.84DD522 pKa = 3.54AITPVFLEE530 pKa = 4.3SFFTINKK537 pKa = 9.68DD538 pKa = 3.41DD539 pKa = 4.66LKK541 pKa = 10.68PLLSRR546 pKa = 11.84MVNSFNKK553 pKa = 10.25YY554 pKa = 7.53SDD556 pKa = 2.61KK557 pKa = 11.28GYY559 pKa = 10.37FVNNFIPLKK568 pKa = 9.87TYY570 pKa = 10.96YY571 pKa = 10.05EE572 pKa = 4.0YY573 pKa = 11.05SAYY576 pKa = 9.84KK577 pKa = 9.44DD578 pKa = 3.64TIKK581 pKa = 11.14SLVCKK586 pKa = 9.31TADD589 pKa = 3.3KK590 pKa = 11.0QQ591 pKa = 3.61
MM1 pKa = 7.43SLVKK5 pKa = 10.64LQFANILLPLEE16 pKa = 4.49VSTSEE21 pKa = 3.55MDD23 pKa = 3.35QFKK26 pKa = 11.44LEE28 pKa = 4.8LINLLMTTFGSVNLFSSFTIFAVLSNNMKK57 pKa = 10.25LHH59 pKa = 6.75PLQLSIVDD67 pKa = 3.61HH68 pKa = 6.2TEE70 pKa = 3.91PGSIPMNRR78 pKa = 11.84EE79 pKa = 3.26VAEE82 pKa = 5.01FIAKK86 pKa = 8.32TFSDD90 pKa = 4.82RR91 pKa = 11.84ICRR94 pKa = 11.84TIGYY98 pKa = 9.34NIYY101 pKa = 8.93TLEE104 pKa = 4.23LAHH107 pKa = 7.15APINNQVDD115 pKa = 3.71DD116 pKa = 4.94DD117 pKa = 4.24FLSRR121 pKa = 11.84LSKK124 pKa = 11.06LNLSSSSSSDD134 pKa = 3.06SGNEE138 pKa = 3.91SFTLTIRR145 pKa = 11.84KK146 pKa = 8.77DD147 pKa = 3.31ASIGQIFDD155 pKa = 4.46SIINNAKK162 pKa = 10.34DD163 pKa = 4.02YY164 pKa = 11.55LPMKK168 pKa = 8.85TIKK171 pKa = 10.36DD172 pKa = 3.54ALKK175 pKa = 10.63ASVDD179 pKa = 3.56KK180 pKa = 10.03FTSPSNASATTTNGSRR196 pKa = 11.84ISRR199 pKa = 11.84SKK201 pKa = 11.18SDD203 pKa = 4.46DD204 pKa = 3.19SLKK207 pKa = 10.19RR208 pKa = 11.84DD209 pKa = 3.61KK210 pKa = 11.17EE211 pKa = 4.21STEE214 pKa = 3.61TDD216 pKa = 2.76KK217 pKa = 11.59RR218 pKa = 11.84MLGNMTASDD227 pKa = 4.65LLNAMSRR234 pKa = 11.84GCSNTCNITCPKK246 pKa = 10.07CEE248 pKa = 4.07YY249 pKa = 10.36IVVKK253 pKa = 10.96NNSTSNNGSITPARR267 pKa = 11.84KK268 pKa = 9.6SNNVSDD274 pKa = 4.26CLFYY278 pKa = 10.69TPQRR282 pKa = 11.84RR283 pKa = 11.84LTDD286 pKa = 3.54YY287 pKa = 7.45EE288 pKa = 4.42TKK290 pKa = 9.84FTVNYY295 pKa = 9.72NSQVYY300 pKa = 8.66NVDD303 pKa = 3.15MVYY306 pKa = 10.24MGMSEE311 pKa = 4.23RR312 pKa = 11.84NKK314 pKa = 9.94TIAFLDD320 pKa = 3.65VLLSLNNRR328 pKa = 11.84RR329 pKa = 11.84LFTNATEE336 pKa = 3.81NSSYY340 pKa = 10.81IKK342 pKa = 10.08RR343 pKa = 11.84HH344 pKa = 4.72YY345 pKa = 9.38KK346 pKa = 10.3RR347 pKa = 11.84NVPVLSTKK355 pKa = 10.42SSNATITTRR364 pKa = 11.84DD365 pKa = 3.67PNSIAMLRR373 pKa = 11.84RR374 pKa = 11.84VTIPPPSTPRR384 pKa = 11.84PLINTFVDD392 pKa = 3.44SFDD395 pKa = 3.49KK396 pKa = 11.68VEE398 pKa = 4.2INPRR402 pKa = 11.84THH404 pKa = 6.96SNIIPYY410 pKa = 9.86PSVNTGVINVRR421 pKa = 11.84NDD423 pKa = 3.49ALKK426 pKa = 10.5KK427 pKa = 10.29IYY429 pKa = 10.07DD430 pKa = 3.86SNGTLIMDD438 pKa = 4.21FTNITLEE445 pKa = 4.28IIRR448 pKa = 11.84EE449 pKa = 3.91GHH451 pKa = 5.06TYY453 pKa = 11.08KK454 pKa = 9.98FTTAKK459 pKa = 10.39GSYY462 pKa = 7.91NTAGEE467 pKa = 4.05FMKK470 pKa = 10.99YY471 pKa = 9.88IVDD474 pKa = 4.09AYY476 pKa = 10.45GLRR479 pKa = 11.84QTLEE483 pKa = 3.74LFMDD487 pKa = 4.62KK488 pKa = 10.52FIDD491 pKa = 3.83MPDD494 pKa = 3.24TDD496 pKa = 4.96FQALLDD502 pKa = 4.63GIIKK506 pKa = 8.89RR507 pKa = 11.84TRR509 pKa = 11.84LMKK512 pKa = 10.66RR513 pKa = 11.84MMHH516 pKa = 6.02NDD518 pKa = 3.04IYY520 pKa = 10.94RR521 pKa = 11.84DD522 pKa = 3.54AITPVFLEE530 pKa = 4.3SFFTINKK537 pKa = 9.68DD538 pKa = 3.41DD539 pKa = 4.66LKK541 pKa = 10.68PLLSRR546 pKa = 11.84MVNSFNKK553 pKa = 10.25YY554 pKa = 7.53SDD556 pKa = 2.61KK557 pKa = 11.28GYY559 pKa = 10.37FVNNFIPLKK568 pKa = 9.87TYY570 pKa = 10.96YY571 pKa = 10.05EE572 pKa = 4.0YY573 pKa = 11.05SAYY576 pKa = 9.84KK577 pKa = 9.44DD578 pKa = 3.64TIKK581 pKa = 11.14SLVCKK586 pKa = 9.31TADD589 pKa = 3.3KK590 pKa = 11.0QQ591 pKa = 3.61
Molecular weight: 66.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1952 |
137 |
1065 |
488.0 |
56.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.996 ± 0.873 | 1.178 ± 0.283 |
6.762 ± 0.302 | 4.918 ± 0.721 |
5.328 ± 0.362 | 3.535 ± 0.579 |
2.049 ± 0.564 | 7.582 ± 0.409 |
7.889 ± 1.052 | 9.939 ± 0.693 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.561 ± 0.432 | 6.506 ± 1.121 |
3.535 ± 0.316 | 2.613 ± 0.46 |
4.355 ± 0.436 | 8.299 ± 0.986 |
6.814 ± 1.36 | 6.148 ± 0.581 |
0.666 ± 0.357 | 5.328 ± 0.577 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
