
Zavarzinia aquatilis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Zavarziniaceae; Zavarzinia
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
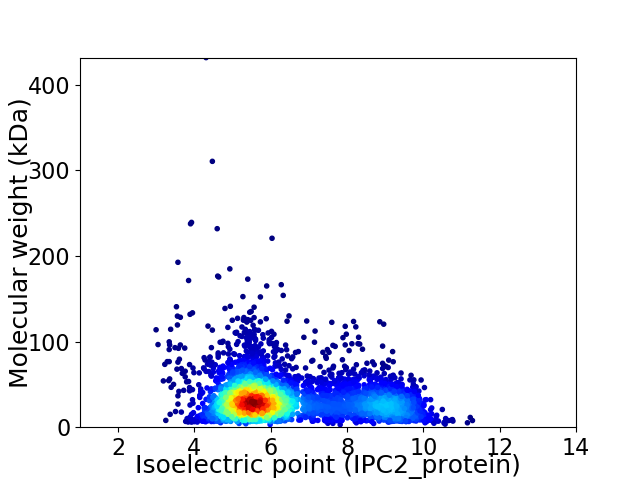
Virtual 2D-PAGE plot for 4178 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317E009|A0A317E009_9PROT Uncharacterized protein OS=Zavarzinia aquatilis OX=2211142 GN=DKG74_15500 PE=3 SV=1
MM1 pKa = 7.46NFTGTNNDD9 pKa = 3.04DD10 pKa = 4.04TYY12 pKa = 11.3NGTSAADD19 pKa = 3.74VFDD22 pKa = 4.0MAQGGVDD29 pKa = 2.92IVNGKK34 pKa = 9.82EE35 pKa = 3.84GDD37 pKa = 3.5DD38 pKa = 3.53EE39 pKa = 4.76FRR41 pKa = 11.84FGATLTAADD50 pKa = 4.45RR51 pKa = 11.84VIGGDD56 pKa = 3.6GYY58 pKa = 11.43DD59 pKa = 3.55HH60 pKa = 7.27VILKK64 pKa = 10.45GDD66 pKa = 3.58YY67 pKa = 10.29SAGLVLSSTTLRR79 pKa = 11.84YY80 pKa = 9.84VEE82 pKa = 4.36SFEE85 pKa = 4.13MTSGFSYY92 pKa = 10.69DD93 pKa = 2.82ITMSNGNLQAGEE105 pKa = 4.22VMTVSAYY112 pKa = 10.2GVGFGGSGHH121 pKa = 5.84YY122 pKa = 9.67FHH124 pKa = 7.55FNGSAEE130 pKa = 3.93TDD132 pKa = 3.06GRR134 pKa = 11.84FNVSDD139 pKa = 4.49GYY141 pKa = 11.64GDD143 pKa = 5.43DD144 pKa = 4.26ILIGSQGGDD153 pKa = 3.93SLSMSYY159 pKa = 10.81GGNDD163 pKa = 3.67YY164 pKa = 10.98IDD166 pKa = 4.01GQGGGDD172 pKa = 3.67VIMAEE177 pKa = 4.29NNFGAGDD184 pKa = 4.65FINGGAGSDD193 pKa = 2.71IVYY196 pKa = 10.1FRR198 pKa = 11.84GLQSATIILNGANFQNIEE216 pKa = 4.01TLAVVGNYY224 pKa = 9.87GADD227 pKa = 3.48FQVADD232 pKa = 4.01TLLAAGQWLNLDD244 pKa = 3.62MRR246 pKa = 11.84SVQAGQTVNFIGSLEE261 pKa = 3.95TDD263 pKa = 3.09GRR265 pKa = 11.84FDD267 pKa = 4.03YY268 pKa = 11.52YY269 pKa = 11.21DD270 pKa = 3.69GAGNDD275 pKa = 3.58HH276 pKa = 6.18FTGGGGADD284 pKa = 4.45LYY286 pKa = 11.03RR287 pKa = 11.84GGNGGSDD294 pKa = 2.89IVSGLGGDD302 pKa = 3.7DD303 pKa = 3.69YY304 pKa = 11.36IYY306 pKa = 10.86FGGDD310 pKa = 3.3LDD312 pKa = 4.93SGDD315 pKa = 4.53TIWGGAGYY323 pKa = 10.68DD324 pKa = 3.9VMDD327 pKa = 3.92LTGDD331 pKa = 4.12LSAGVHH337 pKa = 5.71FADD340 pKa = 5.63DD341 pKa = 3.56GLAGVEE347 pKa = 3.82RR348 pKa = 11.84VVLRR352 pKa = 11.84DD353 pKa = 3.08GFTYY357 pKa = 11.04KK358 pKa = 10.84LFFADD363 pKa = 3.98GNLNTGEE370 pKa = 4.25GLLISSSGNIGAGNSVYY387 pKa = 10.27IDD389 pKa = 3.67ASAEE393 pKa = 3.55TDD395 pKa = 3.2AAYY398 pKa = 9.75TMYY401 pKa = 11.02DD402 pKa = 2.95SDD404 pKa = 5.8GNDD407 pKa = 3.27TLIGGGGNDD416 pKa = 3.95VFWSLAGGRR425 pKa = 11.84DD426 pKa = 3.49KK427 pKa = 11.57LIGNGGNDD435 pKa = 3.53TFVVAGPLHH444 pKa = 7.09AYY446 pKa = 10.2DD447 pKa = 4.68SYY449 pKa = 12.09NGGADD454 pKa = 3.06IDD456 pKa = 4.35TITIYY461 pKa = 10.9NADD464 pKa = 3.69KK465 pKa = 11.24GGVIDD470 pKa = 5.79LSTGKK475 pKa = 10.23ISIGGVAGGSVTQIEE490 pKa = 4.9NISGSAYY497 pKa = 10.55ADD499 pKa = 3.41TLTGNSAANFIDD511 pKa = 4.18GGAGNDD517 pKa = 4.26TISGGQGSDD526 pKa = 3.67DD527 pKa = 3.67LRR529 pKa = 11.84GGAGSDD535 pKa = 3.73KK536 pKa = 10.95LDD538 pKa = 3.98GGAKK542 pKa = 10.13DD543 pKa = 4.64DD544 pKa = 4.53ILYY547 pKa = 10.92GDD549 pKa = 4.71AGNDD553 pKa = 3.1ILLGGQGNDD562 pKa = 3.19NLFGGDD568 pKa = 3.17NTDD571 pKa = 3.99RR572 pKa = 11.84LDD574 pKa = 3.87GGIGNDD580 pKa = 4.36DD581 pKa = 3.59LTGGAGRR588 pKa = 11.84DD589 pKa = 3.29TFVFSTGTGVDD600 pKa = 3.28HH601 pKa = 7.61VKK603 pKa = 10.99DD604 pKa = 3.66FTNNFDD610 pKa = 3.92YY611 pKa = 11.51LDD613 pKa = 3.97FTGVAADD620 pKa = 3.74FADD623 pKa = 3.82IQAHH627 pKa = 5.31MSQVGTDD634 pKa = 3.21VVIAYY639 pKa = 7.49GTDD642 pKa = 3.16KK643 pKa = 11.14FILEE647 pKa = 4.14NTSIGVLDD655 pKa = 4.43ASDD658 pKa = 3.81FLVV661 pKa = 3.42
MM1 pKa = 7.46NFTGTNNDD9 pKa = 3.04DD10 pKa = 4.04TYY12 pKa = 11.3NGTSAADD19 pKa = 3.74VFDD22 pKa = 4.0MAQGGVDD29 pKa = 2.92IVNGKK34 pKa = 9.82EE35 pKa = 3.84GDD37 pKa = 3.5DD38 pKa = 3.53EE39 pKa = 4.76FRR41 pKa = 11.84FGATLTAADD50 pKa = 4.45RR51 pKa = 11.84VIGGDD56 pKa = 3.6GYY58 pKa = 11.43DD59 pKa = 3.55HH60 pKa = 7.27VILKK64 pKa = 10.45GDD66 pKa = 3.58YY67 pKa = 10.29SAGLVLSSTTLRR79 pKa = 11.84YY80 pKa = 9.84VEE82 pKa = 4.36SFEE85 pKa = 4.13MTSGFSYY92 pKa = 10.69DD93 pKa = 2.82ITMSNGNLQAGEE105 pKa = 4.22VMTVSAYY112 pKa = 10.2GVGFGGSGHH121 pKa = 5.84YY122 pKa = 9.67FHH124 pKa = 7.55FNGSAEE130 pKa = 3.93TDD132 pKa = 3.06GRR134 pKa = 11.84FNVSDD139 pKa = 4.49GYY141 pKa = 11.64GDD143 pKa = 5.43DD144 pKa = 4.26ILIGSQGGDD153 pKa = 3.93SLSMSYY159 pKa = 10.81GGNDD163 pKa = 3.67YY164 pKa = 10.98IDD166 pKa = 4.01GQGGGDD172 pKa = 3.67VIMAEE177 pKa = 4.29NNFGAGDD184 pKa = 4.65FINGGAGSDD193 pKa = 2.71IVYY196 pKa = 10.1FRR198 pKa = 11.84GLQSATIILNGANFQNIEE216 pKa = 4.01TLAVVGNYY224 pKa = 9.87GADD227 pKa = 3.48FQVADD232 pKa = 4.01TLLAAGQWLNLDD244 pKa = 3.62MRR246 pKa = 11.84SVQAGQTVNFIGSLEE261 pKa = 3.95TDD263 pKa = 3.09GRR265 pKa = 11.84FDD267 pKa = 4.03YY268 pKa = 11.52YY269 pKa = 11.21DD270 pKa = 3.69GAGNDD275 pKa = 3.58HH276 pKa = 6.18FTGGGGADD284 pKa = 4.45LYY286 pKa = 11.03RR287 pKa = 11.84GGNGGSDD294 pKa = 2.89IVSGLGGDD302 pKa = 3.7DD303 pKa = 3.69YY304 pKa = 11.36IYY306 pKa = 10.86FGGDD310 pKa = 3.3LDD312 pKa = 4.93SGDD315 pKa = 4.53TIWGGAGYY323 pKa = 10.68DD324 pKa = 3.9VMDD327 pKa = 3.92LTGDD331 pKa = 4.12LSAGVHH337 pKa = 5.71FADD340 pKa = 5.63DD341 pKa = 3.56GLAGVEE347 pKa = 3.82RR348 pKa = 11.84VVLRR352 pKa = 11.84DD353 pKa = 3.08GFTYY357 pKa = 11.04KK358 pKa = 10.84LFFADD363 pKa = 3.98GNLNTGEE370 pKa = 4.25GLLISSSGNIGAGNSVYY387 pKa = 10.27IDD389 pKa = 3.67ASAEE393 pKa = 3.55TDD395 pKa = 3.2AAYY398 pKa = 9.75TMYY401 pKa = 11.02DD402 pKa = 2.95SDD404 pKa = 5.8GNDD407 pKa = 3.27TLIGGGGNDD416 pKa = 3.95VFWSLAGGRR425 pKa = 11.84DD426 pKa = 3.49KK427 pKa = 11.57LIGNGGNDD435 pKa = 3.53TFVVAGPLHH444 pKa = 7.09AYY446 pKa = 10.2DD447 pKa = 4.68SYY449 pKa = 12.09NGGADD454 pKa = 3.06IDD456 pKa = 4.35TITIYY461 pKa = 10.9NADD464 pKa = 3.69KK465 pKa = 11.24GGVIDD470 pKa = 5.79LSTGKK475 pKa = 10.23ISIGGVAGGSVTQIEE490 pKa = 4.9NISGSAYY497 pKa = 10.55ADD499 pKa = 3.41TLTGNSAANFIDD511 pKa = 4.18GGAGNDD517 pKa = 4.26TISGGQGSDD526 pKa = 3.67DD527 pKa = 3.67LRR529 pKa = 11.84GGAGSDD535 pKa = 3.73KK536 pKa = 10.95LDD538 pKa = 3.98GGAKK542 pKa = 10.13DD543 pKa = 4.64DD544 pKa = 4.53ILYY547 pKa = 10.92GDD549 pKa = 4.71AGNDD553 pKa = 3.1ILLGGQGNDD562 pKa = 3.19NLFGGDD568 pKa = 3.17NTDD571 pKa = 3.99RR572 pKa = 11.84LDD574 pKa = 3.87GGIGNDD580 pKa = 4.36DD581 pKa = 3.59LTGGAGRR588 pKa = 11.84DD589 pKa = 3.29TFVFSTGTGVDD600 pKa = 3.28HH601 pKa = 7.61VKK603 pKa = 10.99DD604 pKa = 3.66FTNNFDD610 pKa = 3.92YY611 pKa = 11.51LDD613 pKa = 3.97FTGVAADD620 pKa = 3.74FADD623 pKa = 3.82IQAHH627 pKa = 5.31MSQVGTDD634 pKa = 3.21VVIAYY639 pKa = 7.49GTDD642 pKa = 3.16KK643 pKa = 11.14FILEE647 pKa = 4.14NTSIGVLDD655 pKa = 4.43ASDD658 pKa = 3.81FLVV661 pKa = 3.42
Molecular weight: 67.75 kDa
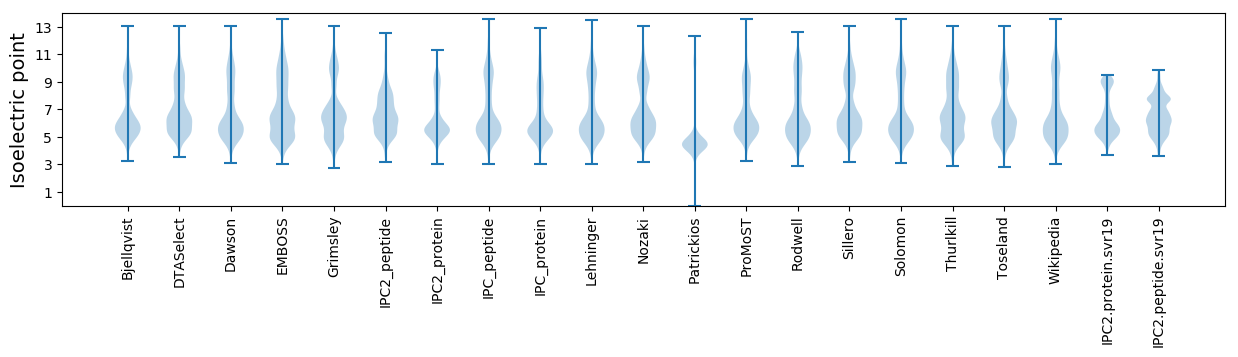
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317EBL6|A0A317EBL6_9PROT Cysteine synthase OS=Zavarzinia aquatilis OX=2211142 GN=cysK PE=4 SV=1
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PSRR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84HH19 pKa = 5.1RR20 pKa = 11.84APARR24 pKa = 11.84PPPRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.32PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PMKK37 pKa = 9.99RR38 pKa = 11.84RR39 pKa = 11.84PASTLPASPPMAPPCWPAARR59 pKa = 11.84RR60 pKa = 11.84ATVRR64 pKa = 3.38
MM1 pKa = 7.47RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PSRR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84SRR18 pKa = 11.84HH19 pKa = 5.1RR20 pKa = 11.84APARR24 pKa = 11.84PPPRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.32PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PMKK37 pKa = 9.99RR38 pKa = 11.84RR39 pKa = 11.84PASTLPASPPMAPPCWPAARR59 pKa = 11.84RR60 pKa = 11.84ATVRR64 pKa = 3.38
Molecular weight: 7.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1369053 |
26 |
4300 |
327.7 |
35.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.587 ± 0.062 | 0.813 ± 0.011 |
6.009 ± 0.036 | 5.376 ± 0.036 |
3.617 ± 0.022 | 9.522 ± 0.057 |
1.931 ± 0.021 | 4.948 ± 0.025 |
2.891 ± 0.028 | 10.412 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.017 | 2.288 ± 0.022 |
5.357 ± 0.039 | 2.684 ± 0.021 |
7.329 ± 0.045 | 4.823 ± 0.028 |
5.197 ± 0.036 | 7.532 ± 0.037 |
1.281 ± 0.017 | 2.136 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
