
Coniella lustricola
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Diaporthales; Schizoparmaceae; Coniella
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
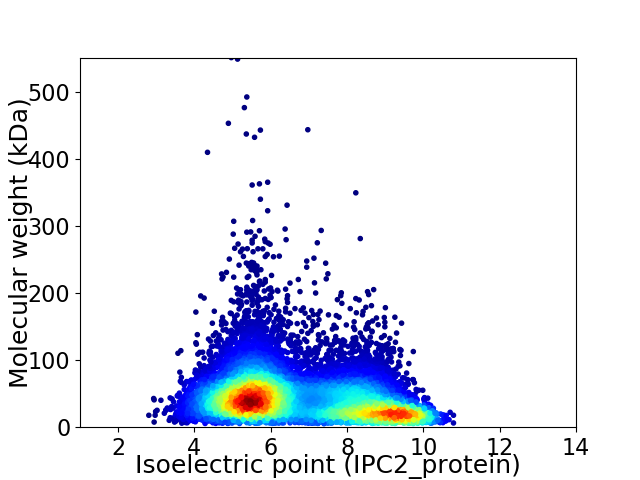
Virtual 2D-PAGE plot for 11309 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T3A7R7|A0A2T3A7R7_9PEZI Uncharacterized protein OS=Coniella lustricola OX=2025994 GN=BD289DRAFT_434149 PE=4 SV=1
MM1 pKa = 7.64KK2 pKa = 9.48FTGSRR7 pKa = 11.84NGLAAAAAALAILQSQVAASPILRR31 pKa = 11.84LHH33 pKa = 6.62RR34 pKa = 11.84KK35 pKa = 8.61SATSIPPFAITPTSSILSSSLTSQPALSMPNGSPAASYY73 pKa = 10.8VAFGLEE79 pKa = 3.89IPITGITGTNPEE91 pKa = 4.13QVFLGSEE98 pKa = 4.19SYY100 pKa = 11.41LLFVQQEE107 pKa = 4.28MTLSADD113 pKa = 3.23VSLNMSNNLLSSSVDD128 pKa = 3.85PIVGFHH134 pKa = 6.74ASSGFSSSSSSVSSSSSSISNSSAGSASDD163 pKa = 4.04LSLLVVLVYY172 pKa = 11.08NNQSTNATSSLNYY185 pKa = 10.2LNQTSQSRR193 pKa = 11.84QFDD196 pKa = 3.67WTSWAASIAADD207 pKa = 3.29SSLRR211 pKa = 11.84MNFDD215 pKa = 3.04MDD217 pKa = 4.17TFASQAGLGKK227 pKa = 10.24AVASEE232 pKa = 4.21FVMITQDD239 pKa = 3.18EE240 pKa = 4.73SSGSGSASYY249 pKa = 10.88SAVTTLSVGAGGASQTAAAVFLTTGLPSAAANGIPTMGAASAAVGVNGMASASLGSGSSAMATGAAAQGGYY320 pKa = 10.64AFASAASGSSATGAAWASSSDD341 pKa = 3.64SSSTSTDD348 pKa = 3.19SNVGVAINNLVSAMSSSMSSIDD370 pKa = 3.79SGVLNSDD377 pKa = 3.23SSDD380 pKa = 3.63NLGSAASGTASMAGGAAVALPTGFASTSLAGSTSSFASSNISSSTSSPTPGTCVAGIYY438 pKa = 9.9KK439 pKa = 10.47CSLDD443 pKa = 3.27GSSYY447 pKa = 8.03QQCASGGTWSVSMAMAAGTTCTPGVSSGLSISASPSTSGSLASSSGSATLSGLSSMDD504 pKa = 3.23SSSDD508 pKa = 3.42SSSSATASGMGLSGLGGLVSLKK530 pKa = 10.53SSQPGSASASGMASEE545 pKa = 5.91GNDD548 pKa = 3.2ADD550 pKa = 4.1IGSSDD555 pKa = 4.05SILPKK560 pKa = 10.61GSFSGASGMGSFEE573 pKa = 4.25SSGNSAFSSGSSTLFHH589 pKa = 6.78HH590 pKa = 7.16GKK592 pKa = 10.36GSSAGAKK599 pKa = 9.76DD600 pKa = 3.43SSSFASSGDD609 pKa = 3.59SGSATSFLSGGSSSSGSGDD628 pKa = 3.18LGSFSASDD636 pKa = 3.89YY637 pKa = 11.05SDD639 pKa = 3.31SHH641 pKa = 7.29PSFLMGGSASGASDD655 pKa = 3.45GLGSLASAGEE665 pKa = 4.35SASSIGSSSSSTQSGGSLSFLGGNSSDD692 pKa = 3.84SQSGDD697 pKa = 3.35SLSSVGVPVTATDD710 pKa = 3.65SSISASSPEE719 pKa = 4.13DD720 pKa = 3.3SCDD723 pKa = 3.42EE724 pKa = 4.62DD725 pKa = 3.93SATTTGSGDD734 pKa = 3.5VSSSTSGSQGYY745 pKa = 9.76DD746 pKa = 3.02SPLSGGISMASGSASSASSASSSFGSSYY774 pKa = 10.42GQSGASASSSTASSLAGSSYY794 pKa = 10.08GQSGTSDD801 pKa = 3.05SSLAASSSSSSAVIPYY817 pKa = 10.1FGGSSASGSVDD828 pKa = 3.44LSGLGDD834 pKa = 3.69STVTDD839 pKa = 3.57SSFGSSASSAYY850 pKa = 10.51GNGASSGMALGGGSSSLSSASSSLGAGSTSVDD882 pKa = 3.4TSSSGASISSVSGDD896 pKa = 3.64SYY898 pKa = 11.39STDD901 pKa = 2.96ASSSGNASSSLSGDD915 pKa = 3.31SSSIDD920 pKa = 3.3ASSSGGDD927 pKa = 3.24ASSSGSASSSLSGDD941 pKa = 3.31SSSIDD946 pKa = 3.3ASSSGGDD953 pKa = 3.24ASSSGSASSSLSGDD967 pKa = 3.31SSSIDD972 pKa = 3.3ASSSGGDD979 pKa = 3.24ASSSGSASSSLSGDD993 pKa = 3.41SSSVDD998 pKa = 3.14ASRR1001 pKa = 11.84TGGDD1005 pKa = 3.46ASSMSGDD1012 pKa = 3.47SSIVDD1017 pKa = 3.61ASSSGGDD1024 pKa = 3.03ASSFGSAMSGASTGSGLEE1042 pKa = 4.42DD1043 pKa = 3.97GSSGGSGLGYY1053 pKa = 10.48LSGSGALGASGTAQAQGAQSAMATGGYY1080 pKa = 9.18QGSAFATAAYY1090 pKa = 9.94QSVASGTASASAAVVTGAASNNLAAALGDD1119 pKa = 3.81ALGNMAVTIGLDD1131 pKa = 2.93IDD1133 pKa = 3.6NGEE1136 pKa = 4.35FEE1138 pKa = 4.56TTITANLKK1146 pKa = 10.48DD1147 pKa = 3.73RR1148 pKa = 11.84TVPTPSPVMWDD1159 pKa = 3.52FRR1161 pKa = 11.84KK1162 pKa = 10.62GSLNARR1168 pKa = 11.84ASPAPSSSSSSSSSSSSTVKK1188 pKa = 10.19SAAAGRR1194 pKa = 11.84VQEE1197 pKa = 4.83AGPAKK1202 pKa = 10.01QVVGVVFALCIASLVMM1218 pKa = 5.16
MM1 pKa = 7.64KK2 pKa = 9.48FTGSRR7 pKa = 11.84NGLAAAAAALAILQSQVAASPILRR31 pKa = 11.84LHH33 pKa = 6.62RR34 pKa = 11.84KK35 pKa = 8.61SATSIPPFAITPTSSILSSSLTSQPALSMPNGSPAASYY73 pKa = 10.8VAFGLEE79 pKa = 3.89IPITGITGTNPEE91 pKa = 4.13QVFLGSEE98 pKa = 4.19SYY100 pKa = 11.41LLFVQQEE107 pKa = 4.28MTLSADD113 pKa = 3.23VSLNMSNNLLSSSVDD128 pKa = 3.85PIVGFHH134 pKa = 6.74ASSGFSSSSSSVSSSSSSISNSSAGSASDD163 pKa = 4.04LSLLVVLVYY172 pKa = 11.08NNQSTNATSSLNYY185 pKa = 10.2LNQTSQSRR193 pKa = 11.84QFDD196 pKa = 3.67WTSWAASIAADD207 pKa = 3.29SSLRR211 pKa = 11.84MNFDD215 pKa = 3.04MDD217 pKa = 4.17TFASQAGLGKK227 pKa = 10.24AVASEE232 pKa = 4.21FVMITQDD239 pKa = 3.18EE240 pKa = 4.73SSGSGSASYY249 pKa = 10.88SAVTTLSVGAGGASQTAAAVFLTTGLPSAAANGIPTMGAASAAVGVNGMASASLGSGSSAMATGAAAQGGYY320 pKa = 10.64AFASAASGSSATGAAWASSSDD341 pKa = 3.64SSSTSTDD348 pKa = 3.19SNVGVAINNLVSAMSSSMSSIDD370 pKa = 3.79SGVLNSDD377 pKa = 3.23SSDD380 pKa = 3.63NLGSAASGTASMAGGAAVALPTGFASTSLAGSTSSFASSNISSSTSSPTPGTCVAGIYY438 pKa = 9.9KK439 pKa = 10.47CSLDD443 pKa = 3.27GSSYY447 pKa = 8.03QQCASGGTWSVSMAMAAGTTCTPGVSSGLSISASPSTSGSLASSSGSATLSGLSSMDD504 pKa = 3.23SSSDD508 pKa = 3.42SSSSATASGMGLSGLGGLVSLKK530 pKa = 10.53SSQPGSASASGMASEE545 pKa = 5.91GNDD548 pKa = 3.2ADD550 pKa = 4.1IGSSDD555 pKa = 4.05SILPKK560 pKa = 10.61GSFSGASGMGSFEE573 pKa = 4.25SSGNSAFSSGSSTLFHH589 pKa = 6.78HH590 pKa = 7.16GKK592 pKa = 10.36GSSAGAKK599 pKa = 9.76DD600 pKa = 3.43SSSFASSGDD609 pKa = 3.59SGSATSFLSGGSSSSGSGDD628 pKa = 3.18LGSFSASDD636 pKa = 3.89YY637 pKa = 11.05SDD639 pKa = 3.31SHH641 pKa = 7.29PSFLMGGSASGASDD655 pKa = 3.45GLGSLASAGEE665 pKa = 4.35SASSIGSSSSSTQSGGSLSFLGGNSSDD692 pKa = 3.84SQSGDD697 pKa = 3.35SLSSVGVPVTATDD710 pKa = 3.65SSISASSPEE719 pKa = 4.13DD720 pKa = 3.3SCDD723 pKa = 3.42EE724 pKa = 4.62DD725 pKa = 3.93SATTTGSGDD734 pKa = 3.5VSSSTSGSQGYY745 pKa = 9.76DD746 pKa = 3.02SPLSGGISMASGSASSASSASSSFGSSYY774 pKa = 10.42GQSGASASSSTASSLAGSSYY794 pKa = 10.08GQSGTSDD801 pKa = 3.05SSLAASSSSSSAVIPYY817 pKa = 10.1FGGSSASGSVDD828 pKa = 3.44LSGLGDD834 pKa = 3.69STVTDD839 pKa = 3.57SSFGSSASSAYY850 pKa = 10.51GNGASSGMALGGGSSSLSSASSSLGAGSTSVDD882 pKa = 3.4TSSSGASISSVSGDD896 pKa = 3.64SYY898 pKa = 11.39STDD901 pKa = 2.96ASSSGNASSSLSGDD915 pKa = 3.31SSSIDD920 pKa = 3.3ASSSGGDD927 pKa = 3.24ASSSGSASSSLSGDD941 pKa = 3.31SSSIDD946 pKa = 3.3ASSSGGDD953 pKa = 3.24ASSSGSASSSLSGDD967 pKa = 3.31SSSIDD972 pKa = 3.3ASSSGGDD979 pKa = 3.24ASSSGSASSSLSGDD993 pKa = 3.41SSSVDD998 pKa = 3.14ASRR1001 pKa = 11.84TGGDD1005 pKa = 3.46ASSMSGDD1012 pKa = 3.47SSIVDD1017 pKa = 3.61ASSSGGDD1024 pKa = 3.03ASSFGSAMSGASTGSGLEE1042 pKa = 4.42DD1043 pKa = 3.97GSSGGSGLGYY1053 pKa = 10.48LSGSGALGASGTAQAQGAQSAMATGGYY1080 pKa = 9.18QGSAFATAAYY1090 pKa = 9.94QSVASGTASASAAVVTGAASNNLAAALGDD1119 pKa = 3.81ALGNMAVTIGLDD1131 pKa = 2.93IDD1133 pKa = 3.6NGEE1136 pKa = 4.35FEE1138 pKa = 4.56TTITANLKK1146 pKa = 10.48DD1147 pKa = 3.73RR1148 pKa = 11.84TVPTPSPVMWDD1159 pKa = 3.52FRR1161 pKa = 11.84KK1162 pKa = 10.62GSLNARR1168 pKa = 11.84ASPAPSSSSSSSSSSSSTVKK1188 pKa = 10.19SAAAGRR1194 pKa = 11.84VQEE1197 pKa = 4.83AGPAKK1202 pKa = 10.01QVVGVVFALCIASLVMM1218 pKa = 5.16
Molecular weight: 114.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T2ZTW6|A0A2T2ZTW6_9PEZI Delta(24)-sterol reductase OS=Coniella lustricola OX=2025994 GN=BD289DRAFT_418248 PE=3 SV=1
MM1 pKa = 7.82AGMLAMLSLVVRR13 pKa = 11.84TLSWRR18 pKa = 11.84PSQTHH23 pKa = 5.28SAAGAAAAGEE33 pKa = 4.62CATKK37 pKa = 8.46TTLSSLHH44 pKa = 5.75GRR46 pKa = 11.84SLRR49 pKa = 11.84RR50 pKa = 11.84SHH52 pKa = 6.16RR53 pKa = 11.84QRR55 pKa = 11.84HH56 pKa = 3.84TRR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84RR64 pKa = 11.84ATAQTVRR71 pKa = 11.84HH72 pKa = 5.76GWAAQQQQPPPQHH85 pKa = 6.68PPRR88 pKa = 11.84PRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84TKK95 pKa = 7.53TTRR98 pKa = 11.84RR99 pKa = 11.84TKK101 pKa = 8.27TTTTSSCASACAAALAAAVAGPAMAAATTPKK132 pKa = 9.67RR133 pKa = 11.84AEE135 pKa = 4.01RR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84GAQSQSATSSRR149 pKa = 11.84TTARR153 pKa = 11.84GRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84CRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84GAGG165 pKa = 3.04
MM1 pKa = 7.82AGMLAMLSLVVRR13 pKa = 11.84TLSWRR18 pKa = 11.84PSQTHH23 pKa = 5.28SAAGAAAAGEE33 pKa = 4.62CATKK37 pKa = 8.46TTLSSLHH44 pKa = 5.75GRR46 pKa = 11.84SLRR49 pKa = 11.84RR50 pKa = 11.84SHH52 pKa = 6.16RR53 pKa = 11.84QRR55 pKa = 11.84HH56 pKa = 3.84TRR58 pKa = 11.84ARR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84RR64 pKa = 11.84ATAQTVRR71 pKa = 11.84HH72 pKa = 5.76GWAAQQQQPPPQHH85 pKa = 6.68PPRR88 pKa = 11.84PRR90 pKa = 11.84RR91 pKa = 11.84PRR93 pKa = 11.84TKK95 pKa = 7.53TTRR98 pKa = 11.84RR99 pKa = 11.84TKK101 pKa = 8.27TTTTSSCASACAAALAAAVAGPAMAAATTPKK132 pKa = 9.67RR133 pKa = 11.84AEE135 pKa = 4.01RR136 pKa = 11.84RR137 pKa = 11.84RR138 pKa = 11.84GAQSQSATSSRR149 pKa = 11.84TTARR153 pKa = 11.84GRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84CRR159 pKa = 11.84RR160 pKa = 11.84RR161 pKa = 11.84RR162 pKa = 11.84GAGG165 pKa = 3.04
Molecular weight: 17.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5000157 |
49 |
4995 |
442.1 |
48.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.45 ± 0.024 | 1.343 ± 0.01 |
5.675 ± 0.016 | 5.716 ± 0.024 |
3.562 ± 0.016 | 6.989 ± 0.021 |
2.517 ± 0.01 | 4.497 ± 0.015 |
4.498 ± 0.019 | 8.828 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.185 ± 0.009 | 3.54 ± 0.012 |
5.997 ± 0.025 | 4.194 ± 0.019 |
6.141 ± 0.019 | 8.448 ± 0.027 |
6.14 ± 0.017 | 6.148 ± 0.016 |
1.479 ± 0.009 | 2.651 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
