
Phytoactinopolyspora halophila
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Jiangellales; Jiangellaceae; Phytoactinopolyspora
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
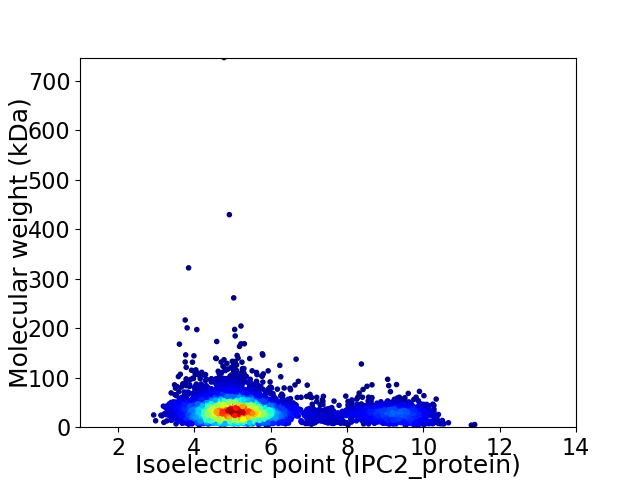
Virtual 2D-PAGE plot for 4405 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A329QS44|A0A329QS44_9ACTN Gamma carbonic anhydrase family protein OS=Phytoactinopolyspora halophila OX=1981511 GN=DPM12_11130 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84RR4 pKa = 11.84LASRR8 pKa = 11.84FSARR12 pKa = 11.84SRR14 pKa = 11.84HH15 pKa = 5.38TDD17 pKa = 3.03RR18 pKa = 11.84QRR20 pKa = 11.84DD21 pKa = 3.56PHH23 pKa = 7.86RR24 pKa = 11.84YY25 pKa = 8.66ASLRR29 pKa = 11.84RR30 pKa = 11.84ASVAAVAATVAGLLVSGAILSAPQGGDD57 pKa = 3.17DD58 pKa = 4.01TADD61 pKa = 3.41STTVRR66 pKa = 11.84VNANGEE72 pKa = 4.23DD73 pKa = 3.96LTDD76 pKa = 3.6STDD79 pKa = 3.6MVWAGDD85 pKa = 3.59QPYY88 pKa = 10.99GSGPWGHH95 pKa = 5.98GTLFGSGSMSGPVTGTDD112 pKa = 3.65DD113 pKa = 4.18DD114 pKa = 4.43EE115 pKa = 6.36LYY117 pKa = 10.83HH118 pKa = 6.87SYY120 pKa = 11.57NLFAAGSGYY129 pKa = 8.89TFDD132 pKa = 5.86LPDD135 pKa = 3.93GDD137 pKa = 4.58YY138 pKa = 10.84EE139 pKa = 4.57VTVKK143 pKa = 9.81TVEE146 pKa = 4.04DD147 pKa = 3.88WATGPGQRR155 pKa = 11.84VFDD158 pKa = 4.0VNVDD162 pKa = 3.61GEE164 pKa = 4.93TVLSEE169 pKa = 3.64FDD171 pKa = 3.35IFAEE175 pKa = 4.7CGASTACDD183 pKa = 3.06RR184 pKa = 11.84SFPASVSGGHH194 pKa = 6.38LSIGFTTNGGSNYY207 pKa = 9.09ATVSAIAITPADD219 pKa = 4.16GGGGDD224 pKa = 4.93PDD226 pKa = 4.18PTPTDD231 pKa = 3.65PTDD234 pKa = 4.25DD235 pKa = 4.11PTPTDD240 pKa = 4.68DD241 pKa = 4.55PTPTDD246 pKa = 4.68DD247 pKa = 4.55PTPTDD252 pKa = 4.61DD253 pKa = 5.15PDD255 pKa = 5.51PGDD258 pKa = 4.08GLPEE262 pKa = 4.31RR263 pKa = 11.84LLTGYY268 pKa = 8.0WHH270 pKa = 6.76NFDD273 pKa = 4.57NNSTVLPLADD283 pKa = 4.02VPDD286 pKa = 4.41SYY288 pKa = 11.98DD289 pKa = 3.38VVAVAFGTEE298 pKa = 4.29DD299 pKa = 3.27VQRR302 pKa = 11.84PGGVDD307 pKa = 3.74FSVSDD312 pKa = 3.98EE313 pKa = 4.32VSDD316 pKa = 4.22SLGGYY321 pKa = 9.75SDD323 pKa = 5.27ADD325 pKa = 3.55LAADD329 pKa = 3.92VEE331 pKa = 4.47QLQEE335 pKa = 3.92RR336 pKa = 11.84GTAVILSIGGEE347 pKa = 4.02RR348 pKa = 11.84GNVRR352 pKa = 11.84VDD354 pKa = 3.3TQEE357 pKa = 4.0RR358 pKa = 11.84ADD360 pKa = 3.83NFADD364 pKa = 3.49SLYY367 pKa = 11.11EE368 pKa = 4.48LMQQYY373 pKa = 10.21GFDD376 pKa = 3.76GVDD379 pKa = 2.85IDD381 pKa = 4.72LEE383 pKa = 4.56HH384 pKa = 7.11GISPAHH390 pKa = 6.35MEE392 pKa = 3.97SALRR396 pKa = 11.84QLADD400 pKa = 3.17RR401 pKa = 11.84VGPEE405 pKa = 4.3LVITMAPQTLDD416 pKa = 3.41MQFTTSDD423 pKa = 3.75YY424 pKa = 11.04FQLALNISDD433 pKa = 3.5ILTVVNTQYY442 pKa = 11.45YY443 pKa = 10.69NSGSMNGCDD452 pKa = 3.39GNVYY456 pKa = 10.5AQGSVEE462 pKa = 5.27FIAALACIQLEE473 pKa = 4.22NGLDD477 pKa = 3.79PDD479 pKa = 3.92QVGLGLPATPSAAGGGYY496 pKa = 10.46VNPTVVNDD504 pKa = 4.74ALDD507 pKa = 3.93CLATGDD513 pKa = 3.68EE514 pKa = 4.98CGAFSPPDD522 pKa = 3.71TYY524 pKa = 11.37PEE526 pKa = 3.62IRR528 pKa = 11.84GAMTWSINWDD538 pKa = 3.1ATNGYY543 pKa = 9.46NFAEE547 pKa = 4.68TVAPHH552 pKa = 6.72LTTLPP557 pKa = 3.64
MM1 pKa = 7.32SRR3 pKa = 11.84RR4 pKa = 11.84LASRR8 pKa = 11.84FSARR12 pKa = 11.84SRR14 pKa = 11.84HH15 pKa = 5.38TDD17 pKa = 3.03RR18 pKa = 11.84QRR20 pKa = 11.84DD21 pKa = 3.56PHH23 pKa = 7.86RR24 pKa = 11.84YY25 pKa = 8.66ASLRR29 pKa = 11.84RR30 pKa = 11.84ASVAAVAATVAGLLVSGAILSAPQGGDD57 pKa = 3.17DD58 pKa = 4.01TADD61 pKa = 3.41STTVRR66 pKa = 11.84VNANGEE72 pKa = 4.23DD73 pKa = 3.96LTDD76 pKa = 3.6STDD79 pKa = 3.6MVWAGDD85 pKa = 3.59QPYY88 pKa = 10.99GSGPWGHH95 pKa = 5.98GTLFGSGSMSGPVTGTDD112 pKa = 3.65DD113 pKa = 4.18DD114 pKa = 4.43EE115 pKa = 6.36LYY117 pKa = 10.83HH118 pKa = 6.87SYY120 pKa = 11.57NLFAAGSGYY129 pKa = 8.89TFDD132 pKa = 5.86LPDD135 pKa = 3.93GDD137 pKa = 4.58YY138 pKa = 10.84EE139 pKa = 4.57VTVKK143 pKa = 9.81TVEE146 pKa = 4.04DD147 pKa = 3.88WATGPGQRR155 pKa = 11.84VFDD158 pKa = 4.0VNVDD162 pKa = 3.61GEE164 pKa = 4.93TVLSEE169 pKa = 3.64FDD171 pKa = 3.35IFAEE175 pKa = 4.7CGASTACDD183 pKa = 3.06RR184 pKa = 11.84SFPASVSGGHH194 pKa = 6.38LSIGFTTNGGSNYY207 pKa = 9.09ATVSAIAITPADD219 pKa = 4.16GGGGDD224 pKa = 4.93PDD226 pKa = 4.18PTPTDD231 pKa = 3.65PTDD234 pKa = 4.25DD235 pKa = 4.11PTPTDD240 pKa = 4.68DD241 pKa = 4.55PTPTDD246 pKa = 4.68DD247 pKa = 4.55PTPTDD252 pKa = 4.61DD253 pKa = 5.15PDD255 pKa = 5.51PGDD258 pKa = 4.08GLPEE262 pKa = 4.31RR263 pKa = 11.84LLTGYY268 pKa = 8.0WHH270 pKa = 6.76NFDD273 pKa = 4.57NNSTVLPLADD283 pKa = 4.02VPDD286 pKa = 4.41SYY288 pKa = 11.98DD289 pKa = 3.38VVAVAFGTEE298 pKa = 4.29DD299 pKa = 3.27VQRR302 pKa = 11.84PGGVDD307 pKa = 3.74FSVSDD312 pKa = 3.98EE313 pKa = 4.32VSDD316 pKa = 4.22SLGGYY321 pKa = 9.75SDD323 pKa = 5.27ADD325 pKa = 3.55LAADD329 pKa = 3.92VEE331 pKa = 4.47QLQEE335 pKa = 3.92RR336 pKa = 11.84GTAVILSIGGEE347 pKa = 4.02RR348 pKa = 11.84GNVRR352 pKa = 11.84VDD354 pKa = 3.3TQEE357 pKa = 4.0RR358 pKa = 11.84ADD360 pKa = 3.83NFADD364 pKa = 3.49SLYY367 pKa = 11.11EE368 pKa = 4.48LMQQYY373 pKa = 10.21GFDD376 pKa = 3.76GVDD379 pKa = 2.85IDD381 pKa = 4.72LEE383 pKa = 4.56HH384 pKa = 7.11GISPAHH390 pKa = 6.35MEE392 pKa = 3.97SALRR396 pKa = 11.84QLADD400 pKa = 3.17RR401 pKa = 11.84VGPEE405 pKa = 4.3LVITMAPQTLDD416 pKa = 3.41MQFTTSDD423 pKa = 3.75YY424 pKa = 11.04FQLALNISDD433 pKa = 3.5ILTVVNTQYY442 pKa = 11.45YY443 pKa = 10.69NSGSMNGCDD452 pKa = 3.39GNVYY456 pKa = 10.5AQGSVEE462 pKa = 5.27FIAALACIQLEE473 pKa = 4.22NGLDD477 pKa = 3.79PDD479 pKa = 3.92QVGLGLPATPSAAGGGYY496 pKa = 10.46VNPTVVNDD504 pKa = 4.74ALDD507 pKa = 3.93CLATGDD513 pKa = 3.68EE514 pKa = 4.98CGAFSPPDD522 pKa = 3.71TYY524 pKa = 11.37PEE526 pKa = 3.62IRR528 pKa = 11.84GAMTWSINWDD538 pKa = 3.1ATNGYY543 pKa = 9.46NFAEE547 pKa = 4.68TVAPHH552 pKa = 6.72LTTLPP557 pKa = 3.64
Molecular weight: 58.65 kDa
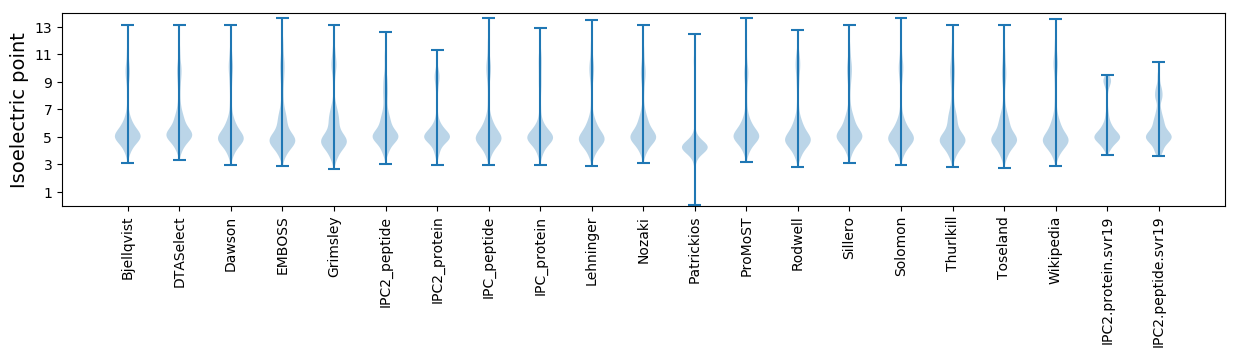
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A329QVR9|A0A329QVR9_9ACTN YbhB/YbcL family Raf kinase inhibitor-like protein OS=Phytoactinopolyspora halophila OX=1981511 GN=DPM12_09960 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84THH16 pKa = 6.18GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84AGRR28 pKa = 11.84AIVSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.73GRR39 pKa = 11.84SKK41 pKa = 10.12LTAA44 pKa = 4.04
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84THH16 pKa = 6.18GFRR19 pKa = 11.84LRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84AGRR28 pKa = 11.84AIVSARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.73GRR39 pKa = 11.84SKK41 pKa = 10.12LTAA44 pKa = 4.04
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1472675 |
25 |
6998 |
334.3 |
36.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.308 ± 0.054 | 0.724 ± 0.009 |
6.849 ± 0.036 | 6.427 ± 0.042 |
2.825 ± 0.023 | 9.035 ± 0.034 |
2.388 ± 0.019 | 4.004 ± 0.023 |
1.56 ± 0.021 | 9.708 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.015 | 1.921 ± 0.018 |
5.621 ± 0.024 | 2.837 ± 0.016 |
7.733 ± 0.039 | 5.555 ± 0.022 |
5.984 ± 0.023 | 8.908 ± 0.031 |
1.527 ± 0.017 | 2.115 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
