
Halomonas anticariensis (strain DSM 16096 / CECT 5854 / LMG 22089 / FP35)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; Halomonas anticariensis
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
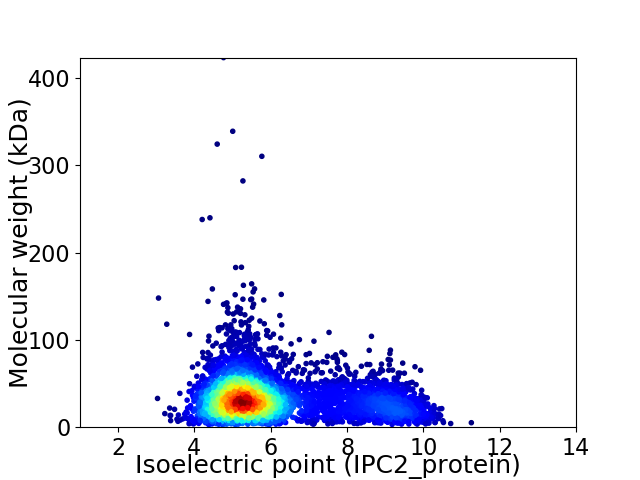
Virtual 2D-PAGE plot for 4705 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S2KNV2|S2KNV2_HALAF Outer-membrane lipoprotein LolB OS=Halomonas anticariensis (strain DSM 16096 / CECT 5854 / LMG 22089 / FP35) OX=1121939 GN=lolB PE=3 SV=1
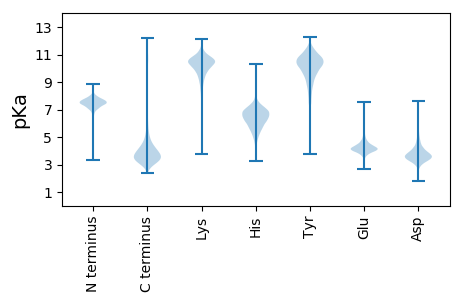
MM1 pKa = 7.0STPSASRR8 pKa = 11.84TLIIPLATLATGLAVGWFAADD29 pKa = 3.29QHH31 pKa = 6.79GDD33 pKa = 3.45SAATDD38 pKa = 3.43ALTNDD43 pKa = 4.11SEE45 pKa = 4.44PLTLEE50 pKa = 3.37EE51 pKa = 4.53TYY53 pKa = 10.61RR54 pKa = 11.84GEE56 pKa = 3.97ITSASALNANDD67 pKa = 3.41GSRR70 pKa = 11.84FEE72 pKa = 4.49RR73 pKa = 11.84LPLALEE79 pKa = 4.53ADD81 pKa = 3.76TLVQLEE87 pKa = 4.61LGGVLNGALALYY99 pKa = 9.98DD100 pKa = 3.63AEE102 pKa = 4.82GNFLAASANDD112 pKa = 5.15GIPVRR117 pKa = 11.84LRR119 pKa = 11.84QRR121 pKa = 11.84IAQDD125 pKa = 3.16GNYY128 pKa = 9.41VLAVSGRR135 pKa = 11.84DD136 pKa = 3.05RR137 pKa = 11.84HH138 pKa = 6.22SYY140 pKa = 10.32GPFRR144 pKa = 11.84ITGHH148 pKa = 5.06TLEE151 pKa = 4.56TQNSGLLTLDD161 pKa = 3.44TPVNGWLQDD170 pKa = 3.2ASNDD174 pKa = 3.72YY175 pKa = 10.61EE176 pKa = 4.6VEE178 pKa = 4.01IPQAGLYY185 pKa = 8.99TIDD188 pKa = 3.71MHH190 pKa = 9.15SDD192 pKa = 3.27DD193 pKa = 4.64LDD195 pKa = 4.99AYY197 pKa = 10.88LVLSGSNGVKK207 pKa = 10.69LEE209 pKa = 4.84DD210 pKa = 4.97DD211 pKa = 4.7DD212 pKa = 5.59SAGDD216 pKa = 3.59LNARR220 pKa = 11.84VSGFLDD226 pKa = 3.3AGRR229 pKa = 11.84YY230 pKa = 6.54QLEE233 pKa = 3.92ARR235 pKa = 11.84TAQGQEE241 pKa = 3.38QGYY244 pKa = 7.76YY245 pKa = 9.03TLEE248 pKa = 4.38LGTRR252 pKa = 11.84DD253 pKa = 3.87LPDD256 pKa = 5.77DD257 pKa = 4.45IEE259 pKa = 4.27LQNGGEE265 pKa = 4.11LALGQPVQGWYY276 pKa = 10.16SGEE279 pKa = 3.82GLEE282 pKa = 4.14YY283 pKa = 10.63RR284 pKa = 11.84LDD286 pKa = 3.46IAEE289 pKa = 4.17QALVTIDD296 pKa = 3.64MQSSEE301 pKa = 3.83FDD303 pKa = 3.22AYY305 pKa = 11.4LEE307 pKa = 4.07LSGNGVMIEE316 pKa = 3.95NDD318 pKa = 3.91DD319 pKa = 3.75GGGNYY324 pKa = 9.81DD325 pKa = 3.91SRR327 pKa = 11.84IQTVLSPGTYY337 pKa = 6.92TVRR340 pKa = 11.84AISYY344 pKa = 8.96DD345 pKa = 3.38SSGSGLFTLEE355 pKa = 4.64AGASDD360 pKa = 3.38IAAAEE365 pKa = 4.08NGVIEE370 pKa = 4.84IGTTVTGQLAGGSQDD385 pKa = 3.31YY386 pKa = 11.31YY387 pKa = 11.17SFHH390 pKa = 7.23VDD392 pKa = 2.45RR393 pKa = 11.84AGQYY397 pKa = 9.54RR398 pKa = 11.84IAMMSSDD405 pKa = 3.26VDD407 pKa = 3.7SYY409 pKa = 11.99LVLQGNGLYY418 pKa = 10.86LEE420 pKa = 5.64DD421 pKa = 5.82DD422 pKa = 4.63DD423 pKa = 6.92GGDD426 pKa = 3.69GYY428 pKa = 11.02NARR431 pKa = 11.84LQTHH435 pKa = 6.86LEE437 pKa = 4.15PGDD440 pKa = 3.69YY441 pKa = 10.85RR442 pKa = 11.84MTARR446 pKa = 11.84TYY448 pKa = 8.14DD449 pKa = 3.45TSGSGSYY456 pKa = 10.16SVSVNSADD464 pKa = 3.87PMLL467 pKa = 4.87
MM1 pKa = 7.0STPSASRR8 pKa = 11.84TLIIPLATLATGLAVGWFAADD29 pKa = 3.29QHH31 pKa = 6.79GDD33 pKa = 3.45SAATDD38 pKa = 3.43ALTNDD43 pKa = 4.11SEE45 pKa = 4.44PLTLEE50 pKa = 3.37EE51 pKa = 4.53TYY53 pKa = 10.61RR54 pKa = 11.84GEE56 pKa = 3.97ITSASALNANDD67 pKa = 3.41GSRR70 pKa = 11.84FEE72 pKa = 4.49RR73 pKa = 11.84LPLALEE79 pKa = 4.53ADD81 pKa = 3.76TLVQLEE87 pKa = 4.61LGGVLNGALALYY99 pKa = 9.98DD100 pKa = 3.63AEE102 pKa = 4.82GNFLAASANDD112 pKa = 5.15GIPVRR117 pKa = 11.84LRR119 pKa = 11.84QRR121 pKa = 11.84IAQDD125 pKa = 3.16GNYY128 pKa = 9.41VLAVSGRR135 pKa = 11.84DD136 pKa = 3.05RR137 pKa = 11.84HH138 pKa = 6.22SYY140 pKa = 10.32GPFRR144 pKa = 11.84ITGHH148 pKa = 5.06TLEE151 pKa = 4.56TQNSGLLTLDD161 pKa = 3.44TPVNGWLQDD170 pKa = 3.2ASNDD174 pKa = 3.72YY175 pKa = 10.61EE176 pKa = 4.6VEE178 pKa = 4.01IPQAGLYY185 pKa = 8.99TIDD188 pKa = 3.71MHH190 pKa = 9.15SDD192 pKa = 3.27DD193 pKa = 4.64LDD195 pKa = 4.99AYY197 pKa = 10.88LVLSGSNGVKK207 pKa = 10.69LEE209 pKa = 4.84DD210 pKa = 4.97DD211 pKa = 4.7DD212 pKa = 5.59SAGDD216 pKa = 3.59LNARR220 pKa = 11.84VSGFLDD226 pKa = 3.3AGRR229 pKa = 11.84YY230 pKa = 6.54QLEE233 pKa = 3.92ARR235 pKa = 11.84TAQGQEE241 pKa = 3.38QGYY244 pKa = 7.76YY245 pKa = 9.03TLEE248 pKa = 4.38LGTRR252 pKa = 11.84DD253 pKa = 3.87LPDD256 pKa = 5.77DD257 pKa = 4.45IEE259 pKa = 4.27LQNGGEE265 pKa = 4.11LALGQPVQGWYY276 pKa = 10.16SGEE279 pKa = 3.82GLEE282 pKa = 4.14YY283 pKa = 10.63RR284 pKa = 11.84LDD286 pKa = 3.46IAEE289 pKa = 4.17QALVTIDD296 pKa = 3.64MQSSEE301 pKa = 3.83FDD303 pKa = 3.22AYY305 pKa = 11.4LEE307 pKa = 4.07LSGNGVMIEE316 pKa = 3.95NDD318 pKa = 3.91DD319 pKa = 3.75GGGNYY324 pKa = 9.81DD325 pKa = 3.91SRR327 pKa = 11.84IQTVLSPGTYY337 pKa = 6.92TVRR340 pKa = 11.84AISYY344 pKa = 8.96DD345 pKa = 3.38SSGSGLFTLEE355 pKa = 4.64AGASDD360 pKa = 3.38IAAAEE365 pKa = 4.08NGVIEE370 pKa = 4.84IGTTVTGQLAGGSQDD385 pKa = 3.31YY386 pKa = 11.31YY387 pKa = 11.17SFHH390 pKa = 7.23VDD392 pKa = 2.45RR393 pKa = 11.84AGQYY397 pKa = 9.54RR398 pKa = 11.84IAMMSSDD405 pKa = 3.26VDD407 pKa = 3.7SYY409 pKa = 11.99LVLQGNGLYY418 pKa = 10.86LEE420 pKa = 5.64DD421 pKa = 5.82DD422 pKa = 4.63DD423 pKa = 6.92GGDD426 pKa = 3.69GYY428 pKa = 11.02NARR431 pKa = 11.84LQTHH435 pKa = 6.86LEE437 pKa = 4.15PGDD440 pKa = 3.69YY441 pKa = 10.85RR442 pKa = 11.84MTARR446 pKa = 11.84TYY448 pKa = 8.14DD449 pKa = 3.45TSGSGSYY456 pKa = 10.16SVSVNSADD464 pKa = 3.87PMLL467 pKa = 4.87
Molecular weight: 49.82 kDa
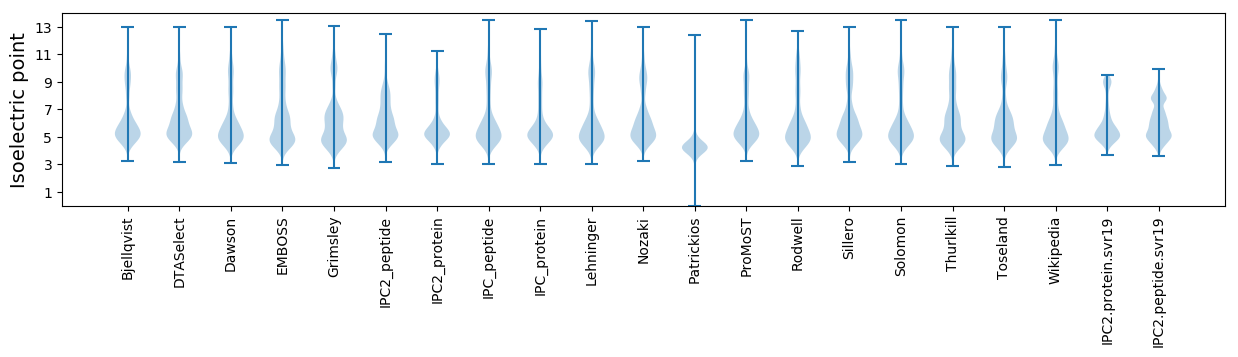
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S2KF15|S2KF15_HALAF Phosphate import ATP-binding protein PstB OS=Halomonas anticariensis (strain DSM 16096 / CECT 5854 / LMG 22089 / FP35) OX=1121939 GN=pstB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1492920 |
29 |
3848 |
317.3 |
34.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.519 ± 0.042 | 0.972 ± 0.013 |
5.739 ± 0.032 | 6.599 ± 0.035 |
3.578 ± 0.023 | 7.998 ± 0.033 |
2.553 ± 0.02 | 5.002 ± 0.034 |
2.795 ± 0.026 | 11.331 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.533 ± 0.017 | 2.682 ± 0.018 |
4.884 ± 0.027 | 3.826 ± 0.027 |
7.108 ± 0.039 | 5.615 ± 0.027 |
5.063 ± 0.024 | 7.188 ± 0.03 |
1.507 ± 0.016 | 2.507 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
