
Cladosporium fulvum T-1 virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Metaviridae; Metavirus
Average proteome isoelectric point is 7.61
Get precalculated fractions of proteins
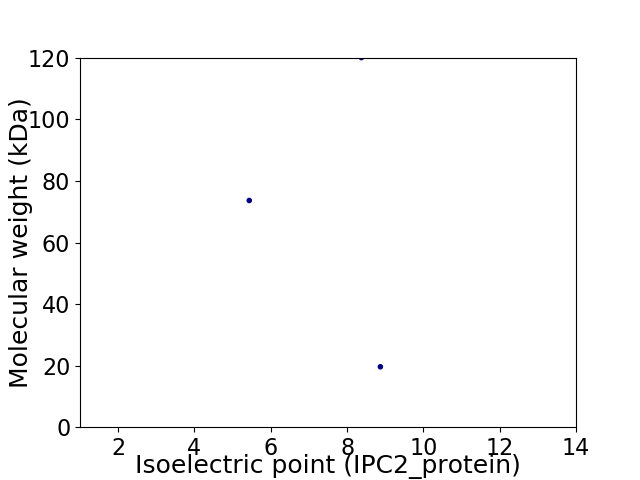
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V9H131|V9H131_9VIRU Env homologue OS=Cladosporium fulvum T-1 virus OX=2052899 PE=4 SV=1
MM1 pKa = 8.17LSMPWLLIQATNPQGWQPEE20 pKa = 4.32FFYY23 pKa = 11.28GDD25 pKa = 3.1RR26 pKa = 11.84VKK28 pKa = 11.0FDD30 pKa = 3.21TWVSQMDD37 pKa = 3.85MYY39 pKa = 11.44FLFNSMTEE47 pKa = 3.81NLKK50 pKa = 10.66PIFATTFLRR59 pKa = 11.84GRR61 pKa = 11.84AQHH64 pKa = 5.21WVKK67 pKa = 10.62PFLRR71 pKa = 11.84KK72 pKa = 9.97YY73 pKa = 10.45LDD75 pKa = 3.84SNGEE79 pKa = 4.06DD80 pKa = 3.39NADD83 pKa = 3.38GVFKK87 pKa = 10.79SYY89 pKa = 10.88NHH91 pKa = 6.87LKK93 pKa = 10.41HH94 pKa = 6.36AMKK97 pKa = 10.57SVFGVSNEE105 pKa = 3.25IATAVRR111 pKa = 11.84VIQHH115 pKa = 6.34LTQKK119 pKa = 9.77TSTAEE124 pKa = 3.83YY125 pKa = 9.06AAKK128 pKa = 9.61FQEE131 pKa = 4.32YY132 pKa = 10.39AQLTDD137 pKa = 3.33WDD139 pKa = 4.52DD140 pKa = 3.43EE141 pKa = 4.44ALQVMYY147 pKa = 10.66RR148 pKa = 11.84RR149 pKa = 11.84GLKK152 pKa = 9.77EE153 pKa = 3.81HH154 pKa = 6.65VKK156 pKa = 10.86DD157 pKa = 3.4EE158 pKa = 4.1LMRR161 pKa = 11.84DD162 pKa = 3.26GRR164 pKa = 11.84KK165 pKa = 8.96IDD167 pKa = 3.52GLGDD171 pKa = 4.0LVQVTIDD178 pKa = 4.76LDD180 pKa = 3.91DD181 pKa = 4.05KK182 pKa = 11.13LYY184 pKa = 10.42EE185 pKa = 3.95RR186 pKa = 11.84AMEE189 pKa = 3.78RR190 pKa = 11.84RR191 pKa = 11.84YY192 pKa = 10.53DD193 pKa = 3.71SKK195 pKa = 11.56VSGKK199 pKa = 10.29AGYY202 pKa = 7.78TPGYY206 pKa = 9.91DD207 pKa = 3.09NRR209 pKa = 11.84NRR211 pKa = 11.84GFNDD215 pKa = 3.18NYY217 pKa = 10.99NKK219 pKa = 10.5PKK221 pKa = 10.45DD222 pKa = 3.6KK223 pKa = 10.2PYY225 pKa = 10.73YY226 pKa = 9.87GPQPMEE232 pKa = 4.14LDD234 pKa = 3.55VTEE237 pKa = 4.57KK238 pKa = 10.54GRR240 pKa = 11.84KK241 pKa = 7.98IRR243 pKa = 11.84NSKK246 pKa = 9.7GNRR249 pKa = 11.84RR250 pKa = 11.84PPSSRR255 pKa = 11.84EE256 pKa = 3.33TRR258 pKa = 11.84TCYY261 pKa = 10.61GCGKK265 pKa = 8.5PGHH268 pKa = 6.71IARR271 pKa = 11.84DD272 pKa = 3.75CRR274 pKa = 11.84GKK276 pKa = 10.62NMVRR280 pKa = 11.84RR281 pKa = 11.84EE282 pKa = 4.3QFNMMQRR289 pKa = 11.84RR290 pKa = 11.84TSKK293 pKa = 10.58SEE295 pKa = 3.86SSVEE299 pKa = 3.92SLGDD303 pKa = 2.86IDD305 pKa = 3.46YY306 pKa = 9.14TRR308 pKa = 11.84RR309 pKa = 11.84VEE311 pKa = 4.03EE312 pKa = 4.03RR313 pKa = 11.84ASNGSTEE320 pKa = 4.1PLLDD324 pKa = 3.99PSQGRR329 pKa = 11.84GGCHH333 pKa = 5.17GQQMVVIPFEE343 pKa = 4.28LQNQPRR349 pKa = 11.84QFNMMARR356 pKa = 11.84RR357 pKa = 11.84VDD359 pKa = 3.66YY360 pKa = 10.52EE361 pKa = 3.9IEE363 pKa = 4.08SEE365 pKa = 4.12SRR367 pKa = 11.84NGHH370 pKa = 6.25GSLHH374 pKa = 6.0WRR376 pKa = 11.84FCYY379 pKa = 9.85QDD381 pKa = 3.16SCQVHH386 pKa = 5.67YY387 pKa = 10.16SAKK390 pKa = 10.24SGAGYY395 pKa = 9.43WPQQPRR401 pKa = 11.84GTLGATHH408 pKa = 6.71RR409 pKa = 11.84QDD411 pKa = 3.04ATLQEE416 pKa = 3.85VDD418 pKa = 3.43IDD420 pKa = 4.13EE421 pKa = 5.01SCFDD425 pKa = 4.93DD426 pKa = 5.8DD427 pKa = 6.69GSDD430 pKa = 5.45KK431 pKa = 11.14EE432 pKa = 4.35NQDD435 pKa = 3.84PGWNGSWSPEE445 pKa = 3.88PQEE448 pKa = 4.48EE449 pKa = 4.43SQEE452 pKa = 4.3TTGLDD457 pKa = 3.74TIEE460 pKa = 4.58EE461 pKa = 4.27DD462 pKa = 3.5QDD464 pKa = 3.76PEE466 pKa = 4.12EE467 pKa = 4.65SSEE470 pKa = 3.97EE471 pKa = 3.92DD472 pKa = 3.12SSEE475 pKa = 4.22GEE477 pKa = 4.34EE478 pKa = 5.53SDD480 pKa = 3.96TDD482 pKa = 4.04DD483 pKa = 6.21DD484 pKa = 4.36NEE486 pKa = 4.23QMTFTVDD493 pKa = 3.36APKK496 pKa = 10.59KK497 pKa = 8.77LYY499 pKa = 11.2DD500 pKa = 4.13MIIHH504 pKa = 6.22LQRR507 pKa = 11.84RR508 pKa = 11.84HH509 pKa = 5.74EE510 pKa = 4.19EE511 pKa = 3.62FLPRR515 pKa = 11.84IGGRR519 pKa = 11.84RR520 pKa = 11.84MLHH523 pKa = 6.16SIEE526 pKa = 4.04FDD528 pKa = 3.1KK529 pKa = 11.43TLDD532 pKa = 3.66TLRR535 pKa = 11.84GMAWGYY541 pKa = 9.9PLMEE545 pKa = 4.29TTEE548 pKa = 4.16NLATAVTEE556 pKa = 4.07RR557 pKa = 11.84PPIGSRR563 pKa = 11.84MIGTGYY569 pKa = 8.29LTPSGTFVSNEE580 pKa = 3.65LRR582 pKa = 11.84NMVQHH587 pKa = 6.69ARR589 pKa = 11.84ALYY592 pKa = 10.68SEE594 pKa = 4.29TQKK597 pKa = 9.91IQEE600 pKa = 4.04RR601 pKa = 11.84HH602 pKa = 4.92RR603 pKa = 11.84VQMWTQKK610 pKa = 10.38GSRR613 pKa = 11.84QVQAEE618 pKa = 4.13ASRR621 pKa = 11.84HH622 pKa = 4.06EE623 pKa = 4.4ARR625 pKa = 11.84RR626 pKa = 11.84STYY629 pKa = 9.34EE630 pKa = 3.56QGKK633 pKa = 10.29GSTPQEE639 pKa = 3.88
MM1 pKa = 8.17LSMPWLLIQATNPQGWQPEE20 pKa = 4.32FFYY23 pKa = 11.28GDD25 pKa = 3.1RR26 pKa = 11.84VKK28 pKa = 11.0FDD30 pKa = 3.21TWVSQMDD37 pKa = 3.85MYY39 pKa = 11.44FLFNSMTEE47 pKa = 3.81NLKK50 pKa = 10.66PIFATTFLRR59 pKa = 11.84GRR61 pKa = 11.84AQHH64 pKa = 5.21WVKK67 pKa = 10.62PFLRR71 pKa = 11.84KK72 pKa = 9.97YY73 pKa = 10.45LDD75 pKa = 3.84SNGEE79 pKa = 4.06DD80 pKa = 3.39NADD83 pKa = 3.38GVFKK87 pKa = 10.79SYY89 pKa = 10.88NHH91 pKa = 6.87LKK93 pKa = 10.41HH94 pKa = 6.36AMKK97 pKa = 10.57SVFGVSNEE105 pKa = 3.25IATAVRR111 pKa = 11.84VIQHH115 pKa = 6.34LTQKK119 pKa = 9.77TSTAEE124 pKa = 3.83YY125 pKa = 9.06AAKK128 pKa = 9.61FQEE131 pKa = 4.32YY132 pKa = 10.39AQLTDD137 pKa = 3.33WDD139 pKa = 4.52DD140 pKa = 3.43EE141 pKa = 4.44ALQVMYY147 pKa = 10.66RR148 pKa = 11.84RR149 pKa = 11.84GLKK152 pKa = 9.77EE153 pKa = 3.81HH154 pKa = 6.65VKK156 pKa = 10.86DD157 pKa = 3.4EE158 pKa = 4.1LMRR161 pKa = 11.84DD162 pKa = 3.26GRR164 pKa = 11.84KK165 pKa = 8.96IDD167 pKa = 3.52GLGDD171 pKa = 4.0LVQVTIDD178 pKa = 4.76LDD180 pKa = 3.91DD181 pKa = 4.05KK182 pKa = 11.13LYY184 pKa = 10.42EE185 pKa = 3.95RR186 pKa = 11.84AMEE189 pKa = 3.78RR190 pKa = 11.84RR191 pKa = 11.84YY192 pKa = 10.53DD193 pKa = 3.71SKK195 pKa = 11.56VSGKK199 pKa = 10.29AGYY202 pKa = 7.78TPGYY206 pKa = 9.91DD207 pKa = 3.09NRR209 pKa = 11.84NRR211 pKa = 11.84GFNDD215 pKa = 3.18NYY217 pKa = 10.99NKK219 pKa = 10.5PKK221 pKa = 10.45DD222 pKa = 3.6KK223 pKa = 10.2PYY225 pKa = 10.73YY226 pKa = 9.87GPQPMEE232 pKa = 4.14LDD234 pKa = 3.55VTEE237 pKa = 4.57KK238 pKa = 10.54GRR240 pKa = 11.84KK241 pKa = 7.98IRR243 pKa = 11.84NSKK246 pKa = 9.7GNRR249 pKa = 11.84RR250 pKa = 11.84PPSSRR255 pKa = 11.84EE256 pKa = 3.33TRR258 pKa = 11.84TCYY261 pKa = 10.61GCGKK265 pKa = 8.5PGHH268 pKa = 6.71IARR271 pKa = 11.84DD272 pKa = 3.75CRR274 pKa = 11.84GKK276 pKa = 10.62NMVRR280 pKa = 11.84RR281 pKa = 11.84EE282 pKa = 4.3QFNMMQRR289 pKa = 11.84RR290 pKa = 11.84TSKK293 pKa = 10.58SEE295 pKa = 3.86SSVEE299 pKa = 3.92SLGDD303 pKa = 2.86IDD305 pKa = 3.46YY306 pKa = 9.14TRR308 pKa = 11.84RR309 pKa = 11.84VEE311 pKa = 4.03EE312 pKa = 4.03RR313 pKa = 11.84ASNGSTEE320 pKa = 4.1PLLDD324 pKa = 3.99PSQGRR329 pKa = 11.84GGCHH333 pKa = 5.17GQQMVVIPFEE343 pKa = 4.28LQNQPRR349 pKa = 11.84QFNMMARR356 pKa = 11.84RR357 pKa = 11.84VDD359 pKa = 3.66YY360 pKa = 10.52EE361 pKa = 3.9IEE363 pKa = 4.08SEE365 pKa = 4.12SRR367 pKa = 11.84NGHH370 pKa = 6.25GSLHH374 pKa = 6.0WRR376 pKa = 11.84FCYY379 pKa = 9.85QDD381 pKa = 3.16SCQVHH386 pKa = 5.67YY387 pKa = 10.16SAKK390 pKa = 10.24SGAGYY395 pKa = 9.43WPQQPRR401 pKa = 11.84GTLGATHH408 pKa = 6.71RR409 pKa = 11.84QDD411 pKa = 3.04ATLQEE416 pKa = 3.85VDD418 pKa = 3.43IDD420 pKa = 4.13EE421 pKa = 5.01SCFDD425 pKa = 4.93DD426 pKa = 5.8DD427 pKa = 6.69GSDD430 pKa = 5.45KK431 pKa = 11.14EE432 pKa = 4.35NQDD435 pKa = 3.84PGWNGSWSPEE445 pKa = 3.88PQEE448 pKa = 4.48EE449 pKa = 4.43SQEE452 pKa = 4.3TTGLDD457 pKa = 3.74TIEE460 pKa = 4.58EE461 pKa = 4.27DD462 pKa = 3.5QDD464 pKa = 3.76PEE466 pKa = 4.12EE467 pKa = 4.65SSEE470 pKa = 3.97EE471 pKa = 3.92DD472 pKa = 3.12SSEE475 pKa = 4.22GEE477 pKa = 4.34EE478 pKa = 5.53SDD480 pKa = 3.96TDD482 pKa = 4.04DD483 pKa = 6.21DD484 pKa = 4.36NEE486 pKa = 4.23QMTFTVDD493 pKa = 3.36APKK496 pKa = 10.59KK497 pKa = 8.77LYY499 pKa = 11.2DD500 pKa = 4.13MIIHH504 pKa = 6.22LQRR507 pKa = 11.84RR508 pKa = 11.84HH509 pKa = 5.74EE510 pKa = 4.19EE511 pKa = 3.62FLPRR515 pKa = 11.84IGGRR519 pKa = 11.84RR520 pKa = 11.84MLHH523 pKa = 6.16SIEE526 pKa = 4.04FDD528 pKa = 3.1KK529 pKa = 11.43TLDD532 pKa = 3.66TLRR535 pKa = 11.84GMAWGYY541 pKa = 9.9PLMEE545 pKa = 4.29TTEE548 pKa = 4.16NLATAVTEE556 pKa = 4.07RR557 pKa = 11.84PPIGSRR563 pKa = 11.84MIGTGYY569 pKa = 8.29LTPSGTFVSNEE580 pKa = 3.65LRR582 pKa = 11.84NMVQHH587 pKa = 6.69ARR589 pKa = 11.84ALYY592 pKa = 10.68SEE594 pKa = 4.29TQKK597 pKa = 9.91IQEE600 pKa = 4.04RR601 pKa = 11.84HH602 pKa = 4.92RR603 pKa = 11.84VQMWTQKK610 pKa = 10.38GSRR613 pKa = 11.84QVQAEE618 pKa = 4.13ASRR621 pKa = 11.84HH622 pKa = 4.06EE623 pKa = 4.4ARR625 pKa = 11.84RR626 pKa = 11.84STYY629 pKa = 9.34EE630 pKa = 3.56QGKK633 pKa = 10.29GSTPQEE639 pKa = 3.88
Molecular weight: 73.65 kDa
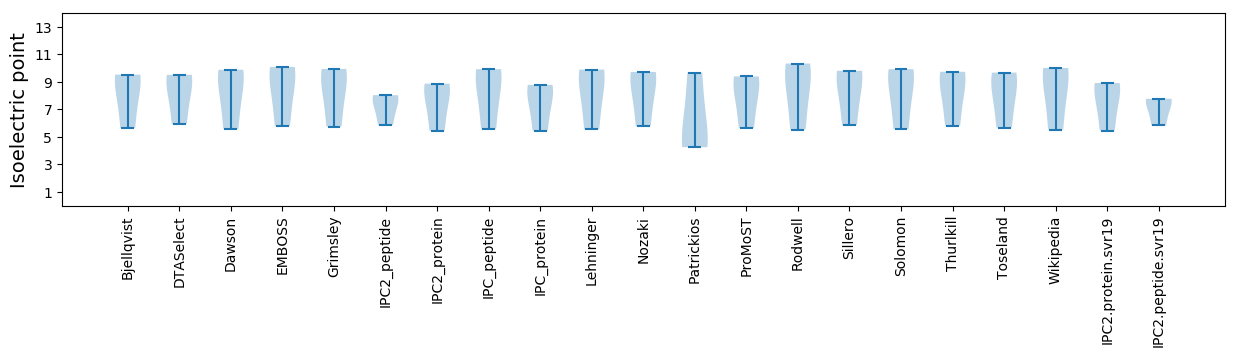
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V9H1L3|V9H1L3_9VIRU Reverse transcriptase (Fragment) OS=Cladosporium fulvum T-1 virus OX=2052899 PE=4 SV=1
MM1 pKa = 7.87APLLKK6 pKa = 10.72EE7 pKa = 3.74GDD9 pKa = 3.63KK10 pKa = 11.25VYY12 pKa = 11.28LLTKK16 pKa = 9.71NLKK19 pKa = 7.89TRR21 pKa = 11.84RR22 pKa = 11.84QTKK25 pKa = 10.16KK26 pKa = 9.92LDD28 pKa = 3.57HH29 pKa = 6.52VKK31 pKa = 10.55VGPFFIDD38 pKa = 3.4KK39 pKa = 10.7VVGPVNYY46 pKa = 10.12RR47 pKa = 11.84LRR49 pKa = 11.84LPPDD53 pKa = 3.52AKK55 pKa = 9.92IHH57 pKa = 5.45PVFHH61 pKa = 6.85ISKK64 pKa = 10.34LEE66 pKa = 4.02PADD69 pKa = 4.78AEE71 pKa = 4.87TPCQEE76 pKa = 4.23SFHH79 pKa = 6.66FEE81 pKa = 3.82PEE83 pKa = 3.89AEE85 pKa = 4.16NEE87 pKa = 4.18FEE89 pKa = 4.64VEE91 pKa = 4.73KK92 pKa = 10.95ILDD95 pKa = 3.83KK96 pKa = 10.96KK97 pKa = 8.32GQRR100 pKa = 11.84YY101 pKa = 7.78LVKK104 pKa = 9.91WKK106 pKa = 10.71GYY108 pKa = 10.27DD109 pKa = 3.23EE110 pKa = 5.74SEE112 pKa = 4.41NTWEE116 pKa = 4.61PRR118 pKa = 11.84INLANCYY125 pKa = 9.51QLLRR129 pKa = 11.84QFQKK133 pKa = 9.63WRR135 pKa = 11.84QDD137 pKa = 2.98SRR139 pKa = 11.84KK140 pKa = 9.57QEE142 pKa = 3.92AQEE145 pKa = 4.02RR146 pKa = 11.84RR147 pKa = 11.84ASPDD151 pKa = 3.04QTRR154 pKa = 11.84SRR156 pKa = 11.84PKK158 pKa = 9.87YY159 pKa = 7.73PHH161 pKa = 6.91ARR163 pKa = 11.84TKK165 pKa = 10.83
MM1 pKa = 7.87APLLKK6 pKa = 10.72EE7 pKa = 3.74GDD9 pKa = 3.63KK10 pKa = 11.25VYY12 pKa = 11.28LLTKK16 pKa = 9.71NLKK19 pKa = 7.89TRR21 pKa = 11.84RR22 pKa = 11.84QTKK25 pKa = 10.16KK26 pKa = 9.92LDD28 pKa = 3.57HH29 pKa = 6.52VKK31 pKa = 10.55VGPFFIDD38 pKa = 3.4KK39 pKa = 10.7VVGPVNYY46 pKa = 10.12RR47 pKa = 11.84LRR49 pKa = 11.84LPPDD53 pKa = 3.52AKK55 pKa = 9.92IHH57 pKa = 5.45PVFHH61 pKa = 6.85ISKK64 pKa = 10.34LEE66 pKa = 4.02PADD69 pKa = 4.78AEE71 pKa = 4.87TPCQEE76 pKa = 4.23SFHH79 pKa = 6.66FEE81 pKa = 3.82PEE83 pKa = 3.89AEE85 pKa = 4.16NEE87 pKa = 4.18FEE89 pKa = 4.64VEE91 pKa = 4.73KK92 pKa = 10.95ILDD95 pKa = 3.83KK96 pKa = 10.96KK97 pKa = 8.32GQRR100 pKa = 11.84YY101 pKa = 7.78LVKK104 pKa = 9.91WKK106 pKa = 10.71GYY108 pKa = 10.27DD109 pKa = 3.23EE110 pKa = 5.74SEE112 pKa = 4.41NTWEE116 pKa = 4.61PRR118 pKa = 11.84INLANCYY125 pKa = 9.51QLLRR129 pKa = 11.84QFQKK133 pKa = 9.63WRR135 pKa = 11.84QDD137 pKa = 2.98SRR139 pKa = 11.84KK140 pKa = 9.57QEE142 pKa = 3.92AQEE145 pKa = 4.02RR146 pKa = 11.84RR147 pKa = 11.84ASPDD151 pKa = 3.04QTRR154 pKa = 11.84SRR156 pKa = 11.84PKK158 pKa = 9.87YY159 pKa = 7.73PHH161 pKa = 6.91ARR163 pKa = 11.84TKK165 pKa = 10.83
Molecular weight: 19.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1849 |
165 |
1045 |
616.3 |
71.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.516 ± 0.452 | 1.19 ± 0.05 |
6.22 ± 0.583 | 7.734 ± 0.567 |
3.245 ± 0.228 | 5.895 ± 0.955 |
3.029 ± 0.2 | 4.543 ± 0.937 |
6.869 ± 1.166 | 7.247 ± 0.666 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.633 | 3.678 ± 0.21 |
5.571 ± 0.565 | 5.192 ± 0.754 |
7.842 ± 0.323 | 6.057 ± 0.665 |
7.68 ± 1.252 | 4.489 ± 0.2 |
1.731 ± 0.018 | 3.678 ± 0.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
