
Sphaerochaeta globosa (strain ATCC BAA-1886 / DSM 22777 / Buddy) (Spirochaeta sp. (strain Buddy))
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Sphaerochaetaceae; Sphaerochaeta; Sphaerochaeta globosa
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
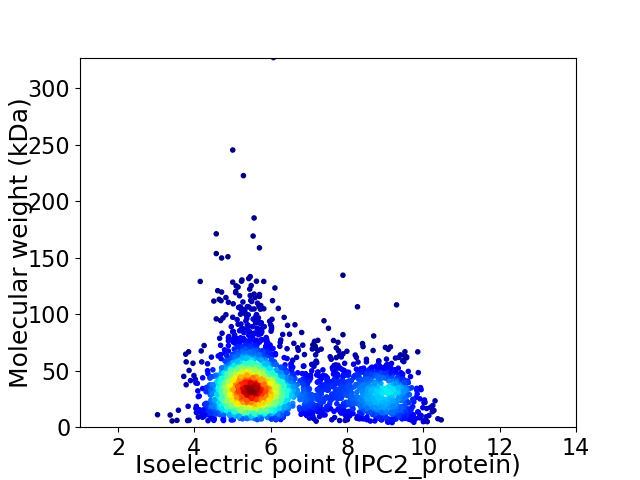
Virtual 2D-PAGE plot for 3007 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
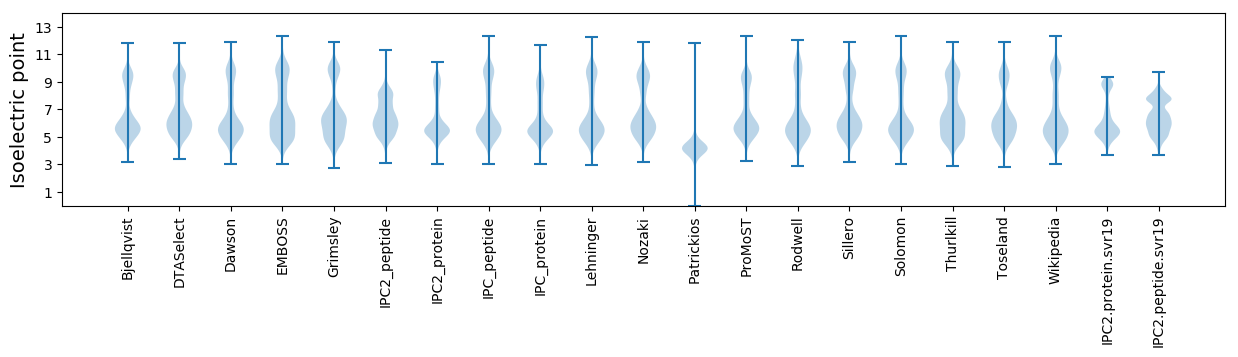
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F0RY60|F0RY60_SPHGB ABC-type transporter integral membrane subunit OS=Sphaerochaeta globosa (strain ATCC BAA-1886 / DSM 22777 / Buddy) OX=158189 GN=SpiBuddy_0732 PE=3 SV=1
MM1 pKa = 7.08NTLKK5 pKa = 10.9NLTKK9 pKa = 10.75YY10 pKa = 10.4VFLFAIVGILSFAVGCSNDD29 pKa = 3.02TTLPINVTGITITGAADD46 pKa = 5.25AITVSNGEE54 pKa = 4.23TLQMSAAVLPLDD66 pKa = 3.97ATDD69 pKa = 5.78ASVTWSVASGSGTATISTTGLLTATAVGTVSVKK102 pKa = 9.76ATANDD107 pKa = 3.34DD108 pKa = 3.62SAYY111 pKa = 10.45EE112 pKa = 4.57GILTITVTAPNVAINIAAITGVTAPVRR139 pKa = 11.84SVTPDD144 pKa = 3.22LTVVEE149 pKa = 4.45TDD151 pKa = 3.17QYY153 pKa = 10.74TATITWAPSDD163 pKa = 3.74SPYY166 pKa = 11.09AGSTVYY172 pKa = 9.78TATITLAPKK181 pKa = 10.3AGYY184 pKa = 6.29TTTGVAADD192 pKa = 3.88AFTVSGADD200 pKa = 3.11TATNLVNSGVVTAVFPATSAAPLARR225 pKa = 11.84VEE227 pKa = 4.69LGTAADD233 pKa = 3.92YY234 pKa = 11.33VILAEE239 pKa = 4.33TLISTTGVTHH249 pKa = 4.74VTGDD253 pKa = 3.85LGISPYY259 pKa = 10.06ATTFITGFALVDD271 pKa = 3.3ATGYY275 pKa = 8.24ATSSLVTGSVYY286 pKa = 10.72AADD289 pKa = 4.11MADD292 pKa = 3.59PTPANLTTAVANMITAYY309 pKa = 10.45NDD311 pKa = 2.94AAARR315 pKa = 11.84PTPDD319 pKa = 3.71EE320 pKa = 4.71LNLGTGAIGGLILAPGLYY338 pKa = 9.75KK339 pKa = 9.85WDD341 pKa = 3.51GTVGIGADD349 pKa = 3.46LTLNGDD355 pKa = 4.56ANDD358 pKa = 2.84IWIFQISGDD367 pKa = 4.06LNMASDD373 pKa = 4.61FEE375 pKa = 4.4VLLTGGAKK383 pKa = 9.92PEE385 pKa = 4.24NIFWQVAGIATLGEE399 pKa = 4.12RR400 pKa = 11.84SHH402 pKa = 6.35MEE404 pKa = 3.84GVILSQTDD412 pKa = 2.94IVLQTGSSINGRR424 pKa = 11.84LLAQTQVTLDD434 pKa = 3.44AATVLEE440 pKa = 4.24PTILL444 pKa = 3.74
MM1 pKa = 7.08NTLKK5 pKa = 10.9NLTKK9 pKa = 10.75YY10 pKa = 10.4VFLFAIVGILSFAVGCSNDD29 pKa = 3.02TTLPINVTGITITGAADD46 pKa = 5.25AITVSNGEE54 pKa = 4.23TLQMSAAVLPLDD66 pKa = 3.97ATDD69 pKa = 5.78ASVTWSVASGSGTATISTTGLLTATAVGTVSVKK102 pKa = 9.76ATANDD107 pKa = 3.34DD108 pKa = 3.62SAYY111 pKa = 10.45EE112 pKa = 4.57GILTITVTAPNVAINIAAITGVTAPVRR139 pKa = 11.84SVTPDD144 pKa = 3.22LTVVEE149 pKa = 4.45TDD151 pKa = 3.17QYY153 pKa = 10.74TATITWAPSDD163 pKa = 3.74SPYY166 pKa = 11.09AGSTVYY172 pKa = 9.78TATITLAPKK181 pKa = 10.3AGYY184 pKa = 6.29TTTGVAADD192 pKa = 3.88AFTVSGADD200 pKa = 3.11TATNLVNSGVVTAVFPATSAAPLARR225 pKa = 11.84VEE227 pKa = 4.69LGTAADD233 pKa = 3.92YY234 pKa = 11.33VILAEE239 pKa = 4.33TLISTTGVTHH249 pKa = 4.74VTGDD253 pKa = 3.85LGISPYY259 pKa = 10.06ATTFITGFALVDD271 pKa = 3.3ATGYY275 pKa = 8.24ATSSLVTGSVYY286 pKa = 10.72AADD289 pKa = 4.11MADD292 pKa = 3.59PTPANLTTAVANMITAYY309 pKa = 10.45NDD311 pKa = 2.94AAARR315 pKa = 11.84PTPDD319 pKa = 3.71EE320 pKa = 4.71LNLGTGAIGGLILAPGLYY338 pKa = 9.75KK339 pKa = 9.85WDD341 pKa = 3.51GTVGIGADD349 pKa = 3.46LTLNGDD355 pKa = 4.56ANDD358 pKa = 2.84IWIFQISGDD367 pKa = 4.06LNMASDD373 pKa = 4.61FEE375 pKa = 4.4VLLTGGAKK383 pKa = 9.92PEE385 pKa = 4.24NIFWQVAGIATLGEE399 pKa = 4.12RR400 pKa = 11.84SHH402 pKa = 6.35MEE404 pKa = 3.84GVILSQTDD412 pKa = 2.94IVLQTGSSINGRR424 pKa = 11.84LLAQTQVTLDD434 pKa = 3.44AATVLEE440 pKa = 4.24PTILL444 pKa = 3.74
Molecular weight: 44.95 kDa
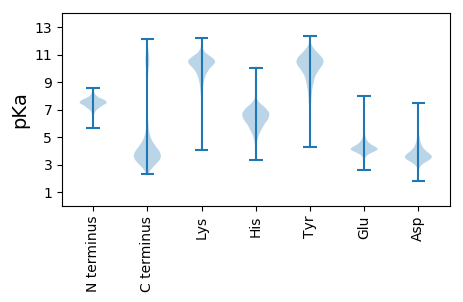
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F0RVE8|F0RVE8_SPHGB Uncharacterized protein OS=Sphaerochaeta globosa (strain ATCC BAA-1886 / DSM 22777 / Buddy) OX=158189 GN=SpiBuddy_1115 PE=4 SV=1
MM1 pKa = 7.08NRR3 pKa = 11.84KK4 pKa = 9.08GLLPNLFIITLSYY17 pKa = 9.99FALGFVSILFANLALLCMVLPFILLAKK44 pKa = 9.91AKK46 pKa = 10.26KK47 pKa = 10.14KK48 pKa = 10.05LWCQTYY54 pKa = 9.98CPRR57 pKa = 11.84ASLLDD62 pKa = 3.71AAGKK66 pKa = 7.25KK67 pKa = 7.98TNWRR71 pKa = 11.84KK72 pKa = 9.57NPSLITSGKK81 pKa = 9.9LRR83 pKa = 11.84GIMLWYY89 pKa = 10.43FGLNLLFITGSTIQVAMLRR108 pKa = 11.84MEE110 pKa = 4.4AMQYY114 pKa = 10.06IRR116 pKa = 11.84LFIAIKK122 pKa = 10.46LFSLPQLITFQAPLFLMHH140 pKa = 7.02LSYY143 pKa = 10.94RR144 pKa = 11.84FYY146 pKa = 11.69GMMASTTILALVLSRR161 pKa = 11.84IYY163 pKa = 10.46RR164 pKa = 11.84PRR166 pKa = 11.84IWCAVCPIGTISDD179 pKa = 3.23ATLRR183 pKa = 11.84VQRR186 pKa = 11.84KK187 pKa = 6.58MKK189 pKa = 10.0PP190 pKa = 3.01
MM1 pKa = 7.08NRR3 pKa = 11.84KK4 pKa = 9.08GLLPNLFIITLSYY17 pKa = 9.99FALGFVSILFANLALLCMVLPFILLAKK44 pKa = 9.91AKK46 pKa = 10.26KK47 pKa = 10.14KK48 pKa = 10.05LWCQTYY54 pKa = 9.98CPRR57 pKa = 11.84ASLLDD62 pKa = 3.71AAGKK66 pKa = 7.25KK67 pKa = 7.98TNWRR71 pKa = 11.84KK72 pKa = 9.57NPSLITSGKK81 pKa = 9.9LRR83 pKa = 11.84GIMLWYY89 pKa = 10.43FGLNLLFITGSTIQVAMLRR108 pKa = 11.84MEE110 pKa = 4.4AMQYY114 pKa = 10.06IRR116 pKa = 11.84LFIAIKK122 pKa = 10.46LFSLPQLITFQAPLFLMHH140 pKa = 7.02LSYY143 pKa = 10.94RR144 pKa = 11.84FYY146 pKa = 11.69GMMASTTILALVLSRR161 pKa = 11.84IYY163 pKa = 10.46RR164 pKa = 11.84PRR166 pKa = 11.84IWCAVCPIGTISDD179 pKa = 3.23ATLRR183 pKa = 11.84VQRR186 pKa = 11.84KK187 pKa = 6.58MKK189 pKa = 10.0PP190 pKa = 3.01
Molecular weight: 21.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003326 |
35 |
2898 |
333.7 |
37.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.064 ± 0.042 | 1.223 ± 0.018 |
5.037 ± 0.035 | 6.103 ± 0.046 |
4.559 ± 0.033 | 6.976 ± 0.037 |
1.963 ± 0.02 | 6.686 ± 0.042 |
5.547 ± 0.037 | 10.931 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.75 ± 0.022 | 3.671 ± 0.028 |
3.781 ± 0.023 | 3.934 ± 0.029 |
4.525 ± 0.034 | 6.977 ± 0.04 |
5.668 ± 0.035 | 6.94 ± 0.038 |
1.035 ± 0.017 | 3.629 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
