
Lynx pardinus (Iberian lynx) (Felis pardina)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria; Boreoeutheria; Laurasiatheria; Carnivora;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
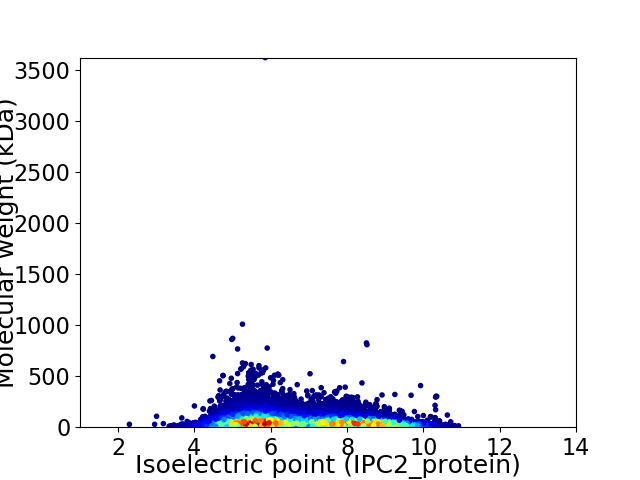
Virtual 2D-PAGE plot for 30929 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
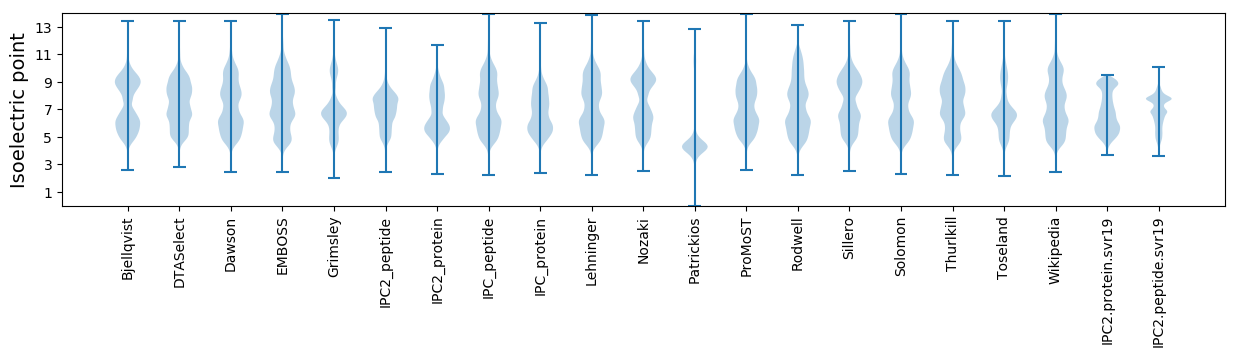
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A485NZD9|A0A485NZD9_LYNPA Isoform of A0A485NYP5 RNA helicase OS=Lynx pardinus OX=191816 GN=LYPA_23C007104 PE=4 SV=1
MM1 pKa = 7.22EE2 pKa = 5.09AVVLIPYY9 pKa = 9.99VLGLLLLPLLAVVLCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.27LPGSYY34 pKa = 10.75DD35 pKa = 3.3NTASDD40 pKa = 3.94SLAPSSIVIKK50 pKa = 10.5RR51 pKa = 11.84PPTLATWTPATSYY64 pKa = 10.76PPVTSYY70 pKa = 11.28PPLSQPDD77 pKa = 4.23LLPIPRR83 pKa = 11.84SPQPPGGSHH92 pKa = 6.82RR93 pKa = 11.84MPSSRR98 pKa = 11.84QDD100 pKa = 3.03SDD102 pKa = 4.67GEE104 pKa = 4.23PACEE108 pKa = 5.61DD109 pKa = 3.61DD110 pKa = 6.68DD111 pKa = 4.6EE112 pKa = 6.88DD113 pKa = 5.49EE114 pKa = 4.94EE115 pKa = 4.44EE116 pKa = 4.91DD117 pKa = 4.15YY118 pKa = 11.46PNEE121 pKa = 4.39GYY123 pKa = 11.04LEE125 pKa = 4.18VLPDD129 pKa = 3.57STPATGTAVPPAPAPSNPGLRR150 pKa = 11.84DD151 pKa = 3.14SAFSMEE157 pKa = 4.23SGEE160 pKa = 4.99DD161 pKa = 3.63YY162 pKa = 11.66VNVPEE167 pKa = 4.61SEE169 pKa = 4.19EE170 pKa = 4.19SADD173 pKa = 3.6VSLDD177 pKa = 3.29GSRR180 pKa = 11.84EE181 pKa = 3.96YY182 pKa = 11.72VNVSQEE188 pKa = 4.24LPPVARR194 pKa = 11.84TEE196 pKa = 3.93PAILSSQNDD205 pKa = 3.92DD206 pKa = 4.02EE207 pKa = 5.46EE208 pKa = 5.21EE209 pKa = 4.2GAPDD213 pKa = 3.75YY214 pKa = 11.56EE215 pKa = 4.31NLQGLNN221 pKa = 3.55
MM1 pKa = 7.22EE2 pKa = 5.09AVVLIPYY9 pKa = 9.99VLGLLLLPLLAVVLCVRR26 pKa = 11.84CRR28 pKa = 11.84EE29 pKa = 4.27LPGSYY34 pKa = 10.75DD35 pKa = 3.3NTASDD40 pKa = 3.94SLAPSSIVIKK50 pKa = 10.5RR51 pKa = 11.84PPTLATWTPATSYY64 pKa = 10.76PPVTSYY70 pKa = 11.28PPLSQPDD77 pKa = 4.23LLPIPRR83 pKa = 11.84SPQPPGGSHH92 pKa = 6.82RR93 pKa = 11.84MPSSRR98 pKa = 11.84QDD100 pKa = 3.03SDD102 pKa = 4.67GEE104 pKa = 4.23PACEE108 pKa = 5.61DD109 pKa = 3.61DD110 pKa = 6.68DD111 pKa = 4.6EE112 pKa = 6.88DD113 pKa = 5.49EE114 pKa = 4.94EE115 pKa = 4.44EE116 pKa = 4.91DD117 pKa = 4.15YY118 pKa = 11.46PNEE121 pKa = 4.39GYY123 pKa = 11.04LEE125 pKa = 4.18VLPDD129 pKa = 3.57STPATGTAVPPAPAPSNPGLRR150 pKa = 11.84DD151 pKa = 3.14SAFSMEE157 pKa = 4.23SGEE160 pKa = 4.99DD161 pKa = 3.63YY162 pKa = 11.66VNVPEE167 pKa = 4.61SEE169 pKa = 4.19EE170 pKa = 4.19SADD173 pKa = 3.6VSLDD177 pKa = 3.29GSRR180 pKa = 11.84EE181 pKa = 3.96YY182 pKa = 11.72VNVSQEE188 pKa = 4.24LPPVARR194 pKa = 11.84TEE196 pKa = 3.93PAILSSQNDD205 pKa = 3.92DD206 pKa = 4.02EE207 pKa = 5.46EE208 pKa = 5.21EE209 pKa = 4.2GAPDD213 pKa = 3.75YY214 pKa = 11.56EE215 pKa = 4.31NLQGLNN221 pKa = 3.55
Molecular weight: 23.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A485NZ17|A0A485NZ17_LYNPA Zinc finger protein 764-like OS=Lynx pardinus OX=191816 GN=LYPA_23C020337 PE=4 SV=1
MM1 pKa = 7.18SAEE4 pKa = 4.25IVWGRR9 pKa = 11.84GRR11 pKa = 11.84TAARR15 pKa = 11.84RR16 pKa = 11.84AAVGGGRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84AAPPTPVRR37 pKa = 11.84GLAGGRR43 pKa = 11.84RR44 pKa = 11.84PGRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84SFSS53 pKa = 3.37
MM1 pKa = 7.18SAEE4 pKa = 4.25IVWGRR9 pKa = 11.84GRR11 pKa = 11.84TAARR15 pKa = 11.84RR16 pKa = 11.84AAVGGGRR23 pKa = 11.84RR24 pKa = 11.84GRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84AAPPTPVRR37 pKa = 11.84GLAGGRR43 pKa = 11.84RR44 pKa = 11.84PGRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84SFSS53 pKa = 3.37
Molecular weight: 5.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17819806 |
42 |
32646 |
576.2 |
63.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.934 ± 0.015 | 2.145 ± 0.01 |
4.802 ± 0.01 | 7.164 ± 0.019 |
3.549 ± 0.011 | 6.504 ± 0.017 |
2.589 ± 0.008 | 4.289 ± 0.013 |
5.806 ± 0.018 | 9.808 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.125 ± 0.007 | 3.571 ± 0.01 |
6.367 ± 0.021 | 4.784 ± 0.014 |
5.716 ± 0.014 | 8.399 ± 0.02 |
5.325 ± 0.013 | 6.019 ± 0.012 |
1.161 ± 0.004 | 2.566 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
