
Cellulosilyticum lentocellum (strain ATCC 49066 / DSM 5427 / NCIMB 11756 / RHM5) (Clostridium lentocellum)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Cellulosilyticum; Cellulosilyticum lentocellum
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
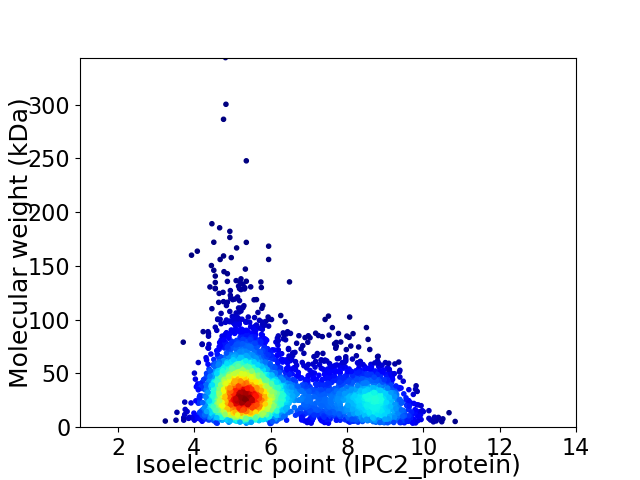
Virtual 2D-PAGE plot for 4150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
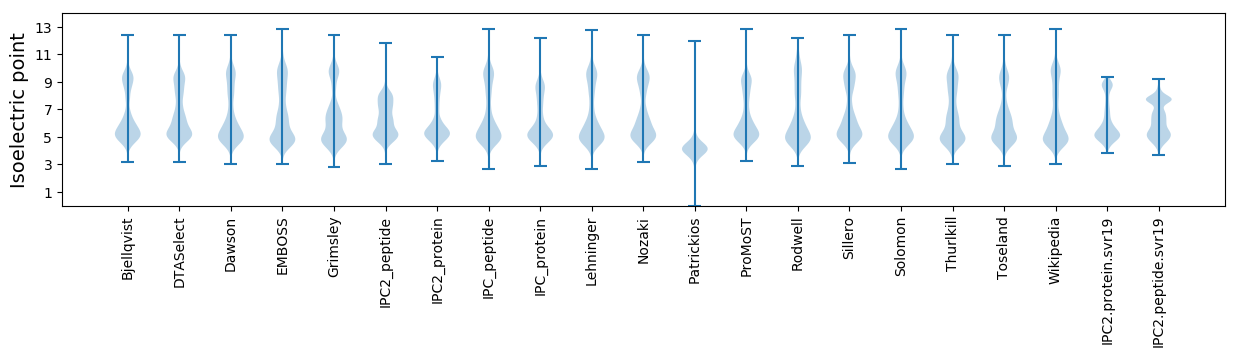
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2JPC1|F2JPC1_CELLD Recombinase OS=Cellulosilyticum lentocellum (strain ATCC 49066 / DSM 5427 / NCIMB 11756 / RHM5) OX=642492 GN=Clole_3165 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 10.39KK3 pKa = 10.25RR4 pKa = 11.84ILSILLCFCMALSLLPMSALAADD27 pKa = 4.46DD28 pKa = 3.97PAVEE32 pKa = 4.16IGGTAVEE39 pKa = 4.27TGDD42 pKa = 3.29WYY44 pKa = 11.1ALTDD48 pKa = 3.53EE49 pKa = 4.97SGTVSTADD57 pKa = 3.41ANADD61 pKa = 2.94NWNVHH66 pKa = 4.4VTKK69 pKa = 10.54DD70 pKa = 3.62DD71 pKa = 3.42TDD73 pKa = 3.69TTVTVTLKK81 pKa = 10.86DD82 pKa = 3.22AAIANNSYY90 pKa = 8.58STHH93 pKa = 6.7AIDD96 pKa = 4.21VNGYY100 pKa = 9.09DD101 pKa = 4.64LEE103 pKa = 4.9IVLEE107 pKa = 4.42GTSNRR112 pKa = 11.84IGTGTTNDD120 pKa = 2.99NGYY123 pKa = 10.79AVYY126 pKa = 10.91NRR128 pKa = 11.84TDD130 pKa = 3.24GKK132 pKa = 10.75DD133 pKa = 3.08ITISGAGDD141 pKa = 3.43LTLTGYY147 pKa = 10.82YY148 pKa = 10.4GISTNGTGSVAVDD161 pKa = 3.06IDD163 pKa = 3.67GSLTVQSQWQMVSCGGQLNATAEE186 pKa = 4.58SITMNGYY193 pKa = 10.08YY194 pKa = 9.52IQCFGGGVSLSAEE207 pKa = 4.05NGDD210 pKa = 3.81VNISGTGDD218 pKa = 3.24YY219 pKa = 10.32GIKK222 pKa = 10.3VGTDD226 pKa = 2.6IMISVPKK233 pKa = 10.1GAVSISGGAYY243 pKa = 9.99SLYY246 pKa = 10.25TSVGNQISVYY256 pKa = 9.86AKK258 pKa = 10.32NDD260 pKa = 2.98ISLTEE265 pKa = 4.25CVQGGDD271 pKa = 3.84GNEE274 pKa = 4.19SQISLTSQTGDD285 pKa = 2.6ITVNSEE291 pKa = 4.02DD292 pKa = 3.61YY293 pKa = 10.75QAIAGVKK300 pKa = 8.94TYY302 pKa = 10.78LGLSAPEE309 pKa = 3.79GDD311 pKa = 2.98ITLTAEE317 pKa = 4.16SLGQNVIAGSNSYY330 pKa = 10.73ALNILAGGTLDD341 pKa = 4.68AQGQDD346 pKa = 3.63GIAGFGTAEE355 pKa = 3.8IKK357 pKa = 10.69AEE359 pKa = 4.13KK360 pKa = 10.54VNIAATGNGYY370 pKa = 10.75GMSGEE375 pKa = 4.46DD376 pKa = 3.29VSVTNPEE383 pKa = 4.06GGNCTEE389 pKa = 4.32ISISGGGDD397 pKa = 2.82GRR399 pKa = 11.84NALNTGNLTLKK410 pKa = 10.26ADD412 pKa = 4.14RR413 pKa = 11.84VIVAAASDD421 pKa = 3.6ASSAISAYY429 pKa = 10.48GNVNIGDD436 pKa = 3.74AGMIIGGTIVSNGTTTIADD455 pKa = 3.39GVLQVQASGPDD466 pKa = 3.36ISSSGLDD473 pKa = 3.94LNTLPAYY480 pKa = 8.34STYY483 pKa = 10.85YY484 pKa = 10.4KK485 pKa = 10.84AGDD488 pKa = 3.91GYY490 pKa = 11.85ALFTPAQNDD499 pKa = 3.72TPAVLTLHH507 pKa = 6.15NAEE510 pKa = 4.47ITSSDD515 pKa = 3.48TPLDD519 pKa = 3.92LGADD523 pKa = 4.14TIIKK527 pKa = 10.54LEE529 pKa = 4.06GSNYY533 pKa = 8.45LTNTNTEE540 pKa = 4.06SGVGIDD546 pKa = 5.02AGDD549 pKa = 3.7NSSGLPQSVIIQGGDD564 pKa = 3.17NDD566 pKa = 4.33SLYY569 pKa = 11.11VSAWQCTSSVEE580 pKa = 3.92NLIISGGKK588 pKa = 6.29VTMYY592 pKa = 9.96GSCYY596 pKa = 10.28GITDD600 pKa = 3.76TGNVLLEE607 pKa = 4.3NGAEE611 pKa = 4.16VSATGGEE618 pKa = 4.24YY619 pKa = 10.97GDD621 pKa = 4.06ALSVGFLEE629 pKa = 5.45DD630 pKa = 5.31DD631 pKa = 4.1EE632 pKa = 5.32NEE634 pKa = 4.11SAARR638 pKa = 11.84NLTINDD644 pKa = 3.74GSSLTAYY651 pKa = 9.43GYY653 pKa = 11.6VMIRR657 pKa = 11.84GNLVMSGEE665 pKa = 4.44SSSFTADD672 pKa = 3.16EE673 pKa = 4.08ISFVLGGLIIGSGSLLTVQPGRR695 pKa = 11.84EE696 pKa = 3.85FYY698 pKa = 11.1VPNSTSSVTNEE709 pKa = 3.33GTLLNNGTLLLPYY722 pKa = 9.51EE723 pKa = 4.31YY724 pKa = 10.86NAADD728 pKa = 3.6VAALNITGSGVVKK741 pKa = 9.96LYY743 pKa = 11.55SMGEE747 pKa = 3.89FPEE750 pKa = 4.13DD751 pKa = 3.0DD752 pKa = 3.51RR753 pKa = 11.84YY754 pKa = 10.82KK755 pKa = 10.88VYY757 pKa = 11.37VNGEE761 pKa = 4.25FYY763 pKa = 11.27ADD765 pKa = 3.44GGEE768 pKa = 4.25LNAQGSLDD776 pKa = 3.8LTTPPTEE783 pKa = 3.41ATYY786 pKa = 10.79YY787 pKa = 10.62KK788 pKa = 10.61SSADD792 pKa = 3.46DD793 pKa = 3.8AYY795 pKa = 9.98IIYY798 pKa = 9.23TPATDD803 pKa = 3.66SEE805 pKa = 4.54NAVLTLHH812 pKa = 6.48GMSNTGYY819 pKa = 8.85LQGGIVLPDD828 pKa = 3.28EE829 pKa = 4.62PVTVRR834 pKa = 11.84VEE836 pKa = 4.33GYY838 pKa = 10.72NEE840 pKa = 4.42IIWISVSKK848 pKa = 9.81EE849 pKa = 3.3IHH851 pKa = 5.25ITGSGRR857 pKa = 11.84LIGRR861 pKa = 11.84IEE863 pKa = 4.02NTASSAEE870 pKa = 3.96LTIDD874 pKa = 3.43SGVTASLMYY883 pKa = 9.22STKK886 pKa = 10.29TGDD889 pKa = 3.4VITNTVYY896 pKa = 11.24GDD898 pKa = 3.54YY899 pKa = 9.71STNAVYY905 pKa = 8.72VTAADD910 pKa = 4.21KK911 pKa = 11.01LVLMPGAVLTVNAGYY926 pKa = 11.08GEE928 pKa = 4.71LFFSEE933 pKa = 4.9GASLSDD939 pKa = 3.25MSIGTGASIVNNNYY953 pKa = 9.76IMLPQGTTAAQIASLPLSGTGIVRR977 pKa = 11.84VATAYY982 pKa = 10.41EE983 pKa = 4.58DD984 pKa = 4.04GSPTAWDD991 pKa = 3.9TYY993 pKa = 9.42TNDD996 pKa = 3.16GAAVKK1001 pKa = 10.41QIGDD1005 pKa = 3.78ADD1007 pKa = 3.92SGLDD1011 pKa = 3.55LTSGEE1016 pKa = 4.36HH1017 pKa = 6.01SGKK1020 pKa = 8.4TVEE1023 pKa = 4.16NDD1025 pKa = 3.29GYY1027 pKa = 11.6AWNSDD1032 pKa = 3.46SKK1034 pKa = 10.19TLTLGDD1040 pKa = 3.89AYY1042 pKa = 10.08IPGDD1046 pKa = 3.57LTLPSGTIVDD1056 pKa = 4.13TTDD1059 pKa = 2.82TTIIGGGLGGEE1070 pKa = 5.0AYY1072 pKa = 7.94TALNITLGGPAPLTISGGIRR1092 pKa = 11.84GGTNGDD1098 pKa = 3.29AVTVQGGAQVTVGDD1112 pKa = 5.05DD1113 pKa = 3.17IFLGASGGADD1123 pKa = 3.01GTLTVTGAGTQLNVSSPYY1141 pKa = 10.52GYY1143 pKa = 11.28AVMCDD1148 pKa = 3.49TVNVQSGASMTANGDD1163 pKa = 4.08SIGVEE1168 pKa = 3.7ALTGVNVTGGSTLTTNCDD1186 pKa = 3.13YY1187 pKa = 11.19GVYY1190 pKa = 10.16IIGGKK1195 pKa = 8.28LTVDD1199 pKa = 3.81DD1200 pKa = 4.19TSKK1203 pKa = 11.3LITNATIAPFCIVDD1217 pKa = 3.66STSNKK1222 pKa = 9.07SQSNVLALAGAPSGTVVASVTGTQAKK1248 pKa = 9.28YY1249 pKa = 9.19WSLVATGGTLSVTDD1263 pKa = 4.12EE1264 pKa = 4.52SNTPVTLSGARR1275 pKa = 11.84TGKK1278 pKa = 8.69LTFVKK1283 pKa = 10.43AATSGGNDD1291 pKa = 3.94DD1292 pKa = 5.36NGGGNNNNGGGASGSSTSYY1311 pKa = 10.24TLTFVTNGGTAISGVSKK1328 pKa = 11.08ASGTVVDD1335 pKa = 4.61LSGYY1339 pKa = 9.38KK1340 pKa = 8.39PTRR1343 pKa = 11.84EE1344 pKa = 4.42GYY1346 pKa = 10.69NFDD1349 pKa = 2.62GWYY1352 pKa = 10.52ADD1354 pKa = 3.52TALTTKK1360 pKa = 8.85VTSVTLTKK1368 pKa = 10.16GTTVYY1373 pKa = 11.04AKK1375 pKa = 8.41WTEE1378 pKa = 4.03KK1379 pKa = 9.9TVQSANPFVDD1389 pKa = 3.77VADD1392 pKa = 3.52SAYY1395 pKa = 11.11YY1396 pKa = 10.24HH1397 pKa = 7.32DD1398 pKa = 4.38AVLWATEE1405 pKa = 3.92KK1406 pKa = 11.05GITSGTTGTTFSPDD1420 pKa = 3.68KK1421 pKa = 10.54ICTRR1425 pKa = 11.84AQAVTFLWRR1434 pKa = 11.84AEE1436 pKa = 4.06GSPEE1440 pKa = 3.81PASANCPFTDD1450 pKa = 4.05VSADD1454 pKa = 3.31DD1455 pKa = 4.24YY1456 pKa = 11.41YY1457 pKa = 11.82YY1458 pKa = 11.18KK1459 pKa = 10.84AVLWAVEE1466 pKa = 3.89KK1467 pKa = 11.02GIVKK1471 pKa = 8.47GTSPTTFSPNDD1482 pKa = 3.45TVTRR1486 pKa = 11.84SQSMTFLWRR1495 pKa = 11.84AAGEE1499 pKa = 4.36TTSISANPFADD1510 pKa = 3.42VSKK1513 pKa = 10.89DD1514 pKa = 2.96AYY1516 pKa = 10.84YY1517 pKa = 11.31YY1518 pKa = 11.04NAVLWAVEE1526 pKa = 3.71KK1527 pKa = 10.79DD1528 pKa = 3.31ITKK1531 pKa = 8.85GTSDD1535 pKa = 3.52TAFSPDD1541 pKa = 3.42GGCTRR1546 pKa = 11.84AQIITLLYY1554 pKa = 10.31RR1555 pKa = 11.84YY1556 pKa = 8.29MGKK1559 pKa = 10.1
MM1 pKa = 7.85KK2 pKa = 10.39KK3 pKa = 10.25RR4 pKa = 11.84ILSILLCFCMALSLLPMSALAADD27 pKa = 4.46DD28 pKa = 3.97PAVEE32 pKa = 4.16IGGTAVEE39 pKa = 4.27TGDD42 pKa = 3.29WYY44 pKa = 11.1ALTDD48 pKa = 3.53EE49 pKa = 4.97SGTVSTADD57 pKa = 3.41ANADD61 pKa = 2.94NWNVHH66 pKa = 4.4VTKK69 pKa = 10.54DD70 pKa = 3.62DD71 pKa = 3.42TDD73 pKa = 3.69TTVTVTLKK81 pKa = 10.86DD82 pKa = 3.22AAIANNSYY90 pKa = 8.58STHH93 pKa = 6.7AIDD96 pKa = 4.21VNGYY100 pKa = 9.09DD101 pKa = 4.64LEE103 pKa = 4.9IVLEE107 pKa = 4.42GTSNRR112 pKa = 11.84IGTGTTNDD120 pKa = 2.99NGYY123 pKa = 10.79AVYY126 pKa = 10.91NRR128 pKa = 11.84TDD130 pKa = 3.24GKK132 pKa = 10.75DD133 pKa = 3.08ITISGAGDD141 pKa = 3.43LTLTGYY147 pKa = 10.82YY148 pKa = 10.4GISTNGTGSVAVDD161 pKa = 3.06IDD163 pKa = 3.67GSLTVQSQWQMVSCGGQLNATAEE186 pKa = 4.58SITMNGYY193 pKa = 10.08YY194 pKa = 9.52IQCFGGGVSLSAEE207 pKa = 4.05NGDD210 pKa = 3.81VNISGTGDD218 pKa = 3.24YY219 pKa = 10.32GIKK222 pKa = 10.3VGTDD226 pKa = 2.6IMISVPKK233 pKa = 10.1GAVSISGGAYY243 pKa = 9.99SLYY246 pKa = 10.25TSVGNQISVYY256 pKa = 9.86AKK258 pKa = 10.32NDD260 pKa = 2.98ISLTEE265 pKa = 4.25CVQGGDD271 pKa = 3.84GNEE274 pKa = 4.19SQISLTSQTGDD285 pKa = 2.6ITVNSEE291 pKa = 4.02DD292 pKa = 3.61YY293 pKa = 10.75QAIAGVKK300 pKa = 8.94TYY302 pKa = 10.78LGLSAPEE309 pKa = 3.79GDD311 pKa = 2.98ITLTAEE317 pKa = 4.16SLGQNVIAGSNSYY330 pKa = 10.73ALNILAGGTLDD341 pKa = 4.68AQGQDD346 pKa = 3.63GIAGFGTAEE355 pKa = 3.8IKK357 pKa = 10.69AEE359 pKa = 4.13KK360 pKa = 10.54VNIAATGNGYY370 pKa = 10.75GMSGEE375 pKa = 4.46DD376 pKa = 3.29VSVTNPEE383 pKa = 4.06GGNCTEE389 pKa = 4.32ISISGGGDD397 pKa = 2.82GRR399 pKa = 11.84NALNTGNLTLKK410 pKa = 10.26ADD412 pKa = 4.14RR413 pKa = 11.84VIVAAASDD421 pKa = 3.6ASSAISAYY429 pKa = 10.48GNVNIGDD436 pKa = 3.74AGMIIGGTIVSNGTTTIADD455 pKa = 3.39GVLQVQASGPDD466 pKa = 3.36ISSSGLDD473 pKa = 3.94LNTLPAYY480 pKa = 8.34STYY483 pKa = 10.85YY484 pKa = 10.4KK485 pKa = 10.84AGDD488 pKa = 3.91GYY490 pKa = 11.85ALFTPAQNDD499 pKa = 3.72TPAVLTLHH507 pKa = 6.15NAEE510 pKa = 4.47ITSSDD515 pKa = 3.48TPLDD519 pKa = 3.92LGADD523 pKa = 4.14TIIKK527 pKa = 10.54LEE529 pKa = 4.06GSNYY533 pKa = 8.45LTNTNTEE540 pKa = 4.06SGVGIDD546 pKa = 5.02AGDD549 pKa = 3.7NSSGLPQSVIIQGGDD564 pKa = 3.17NDD566 pKa = 4.33SLYY569 pKa = 11.11VSAWQCTSSVEE580 pKa = 3.92NLIISGGKK588 pKa = 6.29VTMYY592 pKa = 9.96GSCYY596 pKa = 10.28GITDD600 pKa = 3.76TGNVLLEE607 pKa = 4.3NGAEE611 pKa = 4.16VSATGGEE618 pKa = 4.24YY619 pKa = 10.97GDD621 pKa = 4.06ALSVGFLEE629 pKa = 5.45DD630 pKa = 5.31DD631 pKa = 4.1EE632 pKa = 5.32NEE634 pKa = 4.11SAARR638 pKa = 11.84NLTINDD644 pKa = 3.74GSSLTAYY651 pKa = 9.43GYY653 pKa = 11.6VMIRR657 pKa = 11.84GNLVMSGEE665 pKa = 4.44SSSFTADD672 pKa = 3.16EE673 pKa = 4.08ISFVLGGLIIGSGSLLTVQPGRR695 pKa = 11.84EE696 pKa = 3.85FYY698 pKa = 11.1VPNSTSSVTNEE709 pKa = 3.33GTLLNNGTLLLPYY722 pKa = 9.51EE723 pKa = 4.31YY724 pKa = 10.86NAADD728 pKa = 3.6VAALNITGSGVVKK741 pKa = 9.96LYY743 pKa = 11.55SMGEE747 pKa = 3.89FPEE750 pKa = 4.13DD751 pKa = 3.0DD752 pKa = 3.51RR753 pKa = 11.84YY754 pKa = 10.82KK755 pKa = 10.88VYY757 pKa = 11.37VNGEE761 pKa = 4.25FYY763 pKa = 11.27ADD765 pKa = 3.44GGEE768 pKa = 4.25LNAQGSLDD776 pKa = 3.8LTTPPTEE783 pKa = 3.41ATYY786 pKa = 10.79YY787 pKa = 10.62KK788 pKa = 10.61SSADD792 pKa = 3.46DD793 pKa = 3.8AYY795 pKa = 9.98IIYY798 pKa = 9.23TPATDD803 pKa = 3.66SEE805 pKa = 4.54NAVLTLHH812 pKa = 6.48GMSNTGYY819 pKa = 8.85LQGGIVLPDD828 pKa = 3.28EE829 pKa = 4.62PVTVRR834 pKa = 11.84VEE836 pKa = 4.33GYY838 pKa = 10.72NEE840 pKa = 4.42IIWISVSKK848 pKa = 9.81EE849 pKa = 3.3IHH851 pKa = 5.25ITGSGRR857 pKa = 11.84LIGRR861 pKa = 11.84IEE863 pKa = 4.02NTASSAEE870 pKa = 3.96LTIDD874 pKa = 3.43SGVTASLMYY883 pKa = 9.22STKK886 pKa = 10.29TGDD889 pKa = 3.4VITNTVYY896 pKa = 11.24GDD898 pKa = 3.54YY899 pKa = 9.71STNAVYY905 pKa = 8.72VTAADD910 pKa = 4.21KK911 pKa = 11.01LVLMPGAVLTVNAGYY926 pKa = 11.08GEE928 pKa = 4.71LFFSEE933 pKa = 4.9GASLSDD939 pKa = 3.25MSIGTGASIVNNNYY953 pKa = 9.76IMLPQGTTAAQIASLPLSGTGIVRR977 pKa = 11.84VATAYY982 pKa = 10.41EE983 pKa = 4.58DD984 pKa = 4.04GSPTAWDD991 pKa = 3.9TYY993 pKa = 9.42TNDD996 pKa = 3.16GAAVKK1001 pKa = 10.41QIGDD1005 pKa = 3.78ADD1007 pKa = 3.92SGLDD1011 pKa = 3.55LTSGEE1016 pKa = 4.36HH1017 pKa = 6.01SGKK1020 pKa = 8.4TVEE1023 pKa = 4.16NDD1025 pKa = 3.29GYY1027 pKa = 11.6AWNSDD1032 pKa = 3.46SKK1034 pKa = 10.19TLTLGDD1040 pKa = 3.89AYY1042 pKa = 10.08IPGDD1046 pKa = 3.57LTLPSGTIVDD1056 pKa = 4.13TTDD1059 pKa = 2.82TTIIGGGLGGEE1070 pKa = 5.0AYY1072 pKa = 7.94TALNITLGGPAPLTISGGIRR1092 pKa = 11.84GGTNGDD1098 pKa = 3.29AVTVQGGAQVTVGDD1112 pKa = 5.05DD1113 pKa = 3.17IFLGASGGADD1123 pKa = 3.01GTLTVTGAGTQLNVSSPYY1141 pKa = 10.52GYY1143 pKa = 11.28AVMCDD1148 pKa = 3.49TVNVQSGASMTANGDD1163 pKa = 4.08SIGVEE1168 pKa = 3.7ALTGVNVTGGSTLTTNCDD1186 pKa = 3.13YY1187 pKa = 11.19GVYY1190 pKa = 10.16IIGGKK1195 pKa = 8.28LTVDD1199 pKa = 3.81DD1200 pKa = 4.19TSKK1203 pKa = 11.3LITNATIAPFCIVDD1217 pKa = 3.66STSNKK1222 pKa = 9.07SQSNVLALAGAPSGTVVASVTGTQAKK1248 pKa = 9.28YY1249 pKa = 9.19WSLVATGGTLSVTDD1263 pKa = 4.12EE1264 pKa = 4.52SNTPVTLSGARR1275 pKa = 11.84TGKK1278 pKa = 8.69LTFVKK1283 pKa = 10.43AATSGGNDD1291 pKa = 3.94DD1292 pKa = 5.36NGGGNNNNGGGASGSSTSYY1311 pKa = 10.24TLTFVTNGGTAISGVSKK1328 pKa = 11.08ASGTVVDD1335 pKa = 4.61LSGYY1339 pKa = 9.38KK1340 pKa = 8.39PTRR1343 pKa = 11.84EE1344 pKa = 4.42GYY1346 pKa = 10.69NFDD1349 pKa = 2.62GWYY1352 pKa = 10.52ADD1354 pKa = 3.52TALTTKK1360 pKa = 8.85VTSVTLTKK1368 pKa = 10.16GTTVYY1373 pKa = 11.04AKK1375 pKa = 8.41WTEE1378 pKa = 4.03KK1379 pKa = 9.9TVQSANPFVDD1389 pKa = 3.77VADD1392 pKa = 3.52SAYY1395 pKa = 11.11YY1396 pKa = 10.24HH1397 pKa = 7.32DD1398 pKa = 4.38AVLWATEE1405 pKa = 3.92KK1406 pKa = 11.05GITSGTTGTTFSPDD1420 pKa = 3.68KK1421 pKa = 10.54ICTRR1425 pKa = 11.84AQAVTFLWRR1434 pKa = 11.84AEE1436 pKa = 4.06GSPEE1440 pKa = 3.81PASANCPFTDD1450 pKa = 4.05VSADD1454 pKa = 3.31DD1455 pKa = 4.24YY1456 pKa = 11.41YY1457 pKa = 11.82YY1458 pKa = 11.18KK1459 pKa = 10.84AVLWAVEE1466 pKa = 3.89KK1467 pKa = 11.02GIVKK1471 pKa = 8.47GTSPTTFSPNDD1482 pKa = 3.45TVTRR1486 pKa = 11.84SQSMTFLWRR1495 pKa = 11.84AAGEE1499 pKa = 4.36TTSISANPFADD1510 pKa = 3.42VSKK1513 pKa = 10.89DD1514 pKa = 2.96AYY1516 pKa = 10.84YY1517 pKa = 11.31YY1518 pKa = 11.04NAVLWAVEE1526 pKa = 3.71KK1527 pKa = 10.79DD1528 pKa = 3.31ITKK1531 pKa = 8.85GTSDD1535 pKa = 3.52TAFSPDD1541 pKa = 3.42GGCTRR1546 pKa = 11.84AQIITLLYY1554 pKa = 10.31RR1555 pKa = 11.84YY1556 pKa = 8.29MGKK1559 pKa = 10.1
Molecular weight: 160.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2JSJ6|F2JSJ6_CELLD PSP1 domain protein OS=Cellulosilyticum lentocellum (strain ATCC 49066 / DSM 5427 / NCIMB 11756 / RHM5) OX=642492 GN=Clole_0014 PE=4 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.41KK4 pKa = 10.4SMKK7 pKa = 10.03VKK9 pKa = 9.22QQRR12 pKa = 11.84PAKK15 pKa = 10.02FSTQAYY21 pKa = 5.18TRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 6.8GYY33 pKa = 9.03LRR35 pKa = 11.84KK36 pKa = 10.07YY37 pKa = 9.32GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.19GQIPGVKK57 pKa = 9.25KK58 pKa = 10.9ASWW61 pKa = 3.03
MM1 pKa = 7.61AKK3 pKa = 10.41KK4 pKa = 10.4SMKK7 pKa = 10.03VKK9 pKa = 9.22QQRR12 pKa = 11.84PAKK15 pKa = 10.02FSTQAYY21 pKa = 5.18TRR23 pKa = 11.84CRR25 pKa = 11.84ICGRR29 pKa = 11.84PHH31 pKa = 6.8GYY33 pKa = 9.03LRR35 pKa = 11.84KK36 pKa = 10.07YY37 pKa = 9.32GICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 9.84KK50 pKa = 10.19GQIPGVKK57 pKa = 9.25KK58 pKa = 10.9ASWW61 pKa = 3.03
Molecular weight: 7.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1326943 |
30 |
3068 |
319.7 |
35.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.73 ± 0.039 | 1.231 ± 0.014 |
4.989 ± 0.031 | 7.734 ± 0.044 |
4.006 ± 0.025 | 6.666 ± 0.039 |
1.739 ± 0.018 | 8.506 ± 0.043 |
7.595 ± 0.032 | 9.421 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.84 ± 0.018 | 4.857 ± 0.028 |
3.116 ± 0.02 | 3.438 ± 0.02 |
3.493 ± 0.025 | 5.956 ± 0.032 |
5.788 ± 0.034 | 6.667 ± 0.033 |
0.889 ± 0.015 | 4.339 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
