
Mesorhizobium sp. B2-3-3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Mesorhizobium; unclassified Mesorhizobium
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
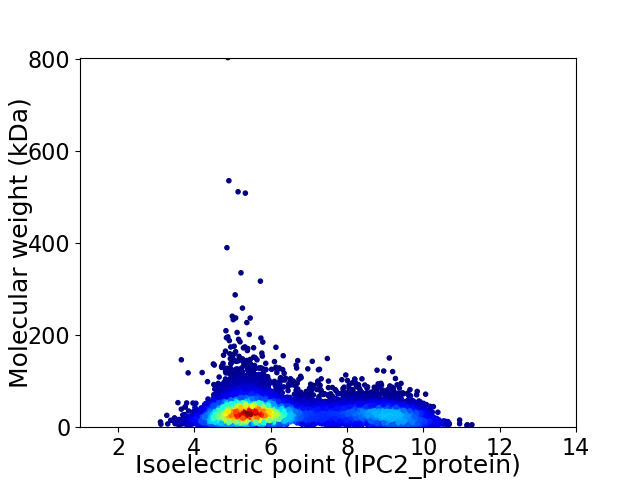
Virtual 2D-PAGE plot for 12366 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A504DPS0|A0A504DPS0_9RHIZ ActR/PrrA/RegA family redox response regulator transcription factor OS=Mesorhizobium sp. B2-3-3 OX=2589960 GN=FKO01_38735 PE=4 SV=1
SS1 pKa = 5.77EE2 pKa = 4.3TTTGSITYY10 pKa = 7.68TAPDD14 pKa = 3.85GPATVTIGGLAVSAVGQTFAGSFGTLTITSIAAGSVGYY52 pKa = 10.45SYY54 pKa = 10.34TLTTNTSGDD63 pKa = 3.54TTHH66 pKa = 7.63DD67 pKa = 3.71DD68 pKa = 3.7FAVSVTDD75 pKa = 4.02ADD77 pKa = 4.39GDD79 pKa = 4.17HH80 pKa = 6.97SDD82 pKa = 3.4KK83 pKa = 11.0TLTINIVDD91 pKa = 4.45DD92 pKa = 4.18VPTAHH97 pKa = 7.6ADD99 pKa = 3.69TDD101 pKa = 4.26SVTAGSYY108 pKa = 10.69GPEE111 pKa = 4.14LGNVITAVGTTSSGADD127 pKa = 3.14TLGADD132 pKa = 3.94GAVVAGVAAGSGGNLDD148 pKa = 3.5NAATVGAVIHH158 pKa = 5.69GTYY161 pKa = 11.02GDD163 pKa = 4.11LTLQANGDD171 pKa = 3.67YY172 pKa = 10.71SYY174 pKa = 11.38ARR176 pKa = 11.84HH177 pKa = 6.44AGTPGGVNDD186 pKa = 3.59VFTYY190 pKa = 8.39TLKK193 pKa = 11.21DD194 pKa = 3.22GDD196 pKa = 4.49GDD198 pKa = 4.17LSHH201 pKa = 6.69TTLTISIGDD210 pKa = 3.77SGVTTTVPVAGTAGAQVSEE229 pKa = 4.85AGLPPHH235 pKa = 6.78GALPAGSGEE244 pKa = 4.44SADD247 pKa = 3.72GLPNNNSDD255 pKa = 3.44TSEE258 pKa = 4.17TTTGSITYY266 pKa = 7.68TAPDD270 pKa = 3.85GPATVTIGGLAVSAVGQTFAGSFGTLTITSIAAGSVGYY308 pKa = 10.45SYY310 pKa = 10.34TLTTNTSGDD319 pKa = 3.54TTHH322 pKa = 7.63DD323 pKa = 3.71DD324 pKa = 3.7FAVSVTDD331 pKa = 4.02ADD333 pKa = 4.39GDD335 pKa = 4.17HH336 pKa = 6.97SDD338 pKa = 3.4KK339 pKa = 11.0TLTINIVDD347 pKa = 4.45DD348 pKa = 4.18VPTAHH353 pKa = 7.26ADD355 pKa = 3.05SWAPAITAPTVLTGLLGNDD374 pKa = 3.07VFGADD379 pKa = 5.05GIATTTPGMVTTTNGAHH396 pKa = 5.83GTVTYY401 pKa = 11.11NNDD404 pKa = 2.79GTFTYY409 pKa = 9.37TPTGIYY415 pKa = 9.86VGSDD419 pKa = 2.42SFTYY423 pKa = 9.66TIKK426 pKa = 11.15DD427 pKa = 3.27GDD429 pKa = 4.42GDD431 pKa = 3.96TSTATVTVNVQTNTVPGGGGTASLTLNEE459 pKa = 4.55AALDD463 pKa = 4.08TTQDD467 pKa = 3.36AADD470 pKa = 3.91LHH472 pKa = 6.89AGAVTGTNPTSPGEE486 pKa = 4.21SAQATSGITFTTTGEE501 pKa = 4.49AITVAFANPTGDD513 pKa = 4.26PNWVAPAVSGLAAGYY528 pKa = 9.49SISWALSGGQLVGTLMQGVTNLGPAIYY555 pKa = 9.75LALSNTSAGANSSLTPVVTATLTDD579 pKa = 3.9QLQHH583 pKa = 6.07TAGSGNITINGLQVVATDD601 pKa = 3.35TSGDD605 pKa = 3.86HH606 pKa = 6.85VSGAVNLTVLDD617 pKa = 4.44DD618 pKa = 4.23APLPFVGDD626 pKa = 4.31SISLPDD632 pKa = 3.88TLHH635 pKa = 6.02TALTEE640 pKa = 4.23HH641 pKa = 7.02LNFAPGAGADD651 pKa = 3.65GVGNVVFNVTTGAAVQDD668 pKa = 3.51ASGANVYY675 pKa = 11.03LNGEE679 pKa = 3.88QLFYY683 pKa = 10.77QVSADD688 pKa = 3.02GHH690 pKa = 4.7TVEE693 pKa = 5.53GRR695 pKa = 11.84SSVANGNDD703 pKa = 3.24LGFTATLNPATDD715 pKa = 2.88TWSFVLNGTIFNGSQFTTAGATATGGNNQVNAFTALTSPDD755 pKa = 3.78TPNDD759 pKa = 3.69LLVTANGSNNVNTSTGNFGVGSGQSITSGEE789 pKa = 4.24TIRR792 pKa = 11.84FDD794 pKa = 3.93FVTNASTTGTIAGTNYY810 pKa = 5.62TTHH813 pKa = 7.09YY814 pKa = 9.0EE815 pKa = 4.2VSSFTQGITKK825 pKa = 10.64ANGTVDD831 pKa = 2.99ITIRR835 pKa = 11.84AVNADD840 pKa = 3.62DD841 pKa = 5.61DD842 pKa = 4.23KK843 pKa = 11.63TFVGDD848 pKa = 3.93GSGEE852 pKa = 3.9ATATGITVTFVNGSGGAAPTLTNNGDD878 pKa = 3.73GTFTLHH884 pKa = 7.45GLDD887 pKa = 3.73QGDD890 pKa = 4.07TFTITASSDD899 pKa = 3.17PFSAVEE905 pKa = 3.65ISGASSGTFKK915 pKa = 11.06LGAASFATANAVDD928 pKa = 4.52PFDD931 pKa = 4.29INIPVTGTDD940 pKa = 3.53GDD942 pKa = 4.08SDD944 pKa = 3.91TAAGSVTAHH953 pKa = 7.55LYY955 pKa = 7.03PTATTLEE962 pKa = 4.28GTNGVDD968 pKa = 3.24NLVATAVKK976 pKa = 9.59TILLGQDD983 pKa = 3.33GDD985 pKa = 4.05DD986 pKa = 4.13HH987 pKa = 6.3LTGINGHH994 pKa = 5.8DD995 pKa = 3.95TILSGGQGNDD1005 pKa = 3.04TLTGLDD1011 pKa = 4.25GKK1013 pKa = 9.72DD1014 pKa = 3.42TLIGGAGADD1023 pKa = 3.76TMTGGAGNDD1032 pKa = 3.53TFVIGAGEE1040 pKa = 4.26STPVIGGSGNAGTISGYY1057 pKa = 10.7DD1058 pKa = 3.2IITDD1062 pKa = 4.14FVAGTDD1068 pKa = 3.47KK1069 pKa = 10.68LTLPGSLVAATSGNVNGGDD1088 pKa = 3.52STLTIGGDD1096 pKa = 3.76TVQSHH1101 pKa = 5.73SVTNGIASFFGTDD1114 pKa = 3.35SFTAPLAVTSAASLAAVVQYY1134 pKa = 11.25LNATDD1139 pKa = 3.82IGNAGATLAFTATISGVNHH1158 pKa = 6.48TYY1160 pKa = 10.79VYY1162 pKa = 9.58TQATTNAGVGALVDD1176 pKa = 4.18LQGVTISNLNTLIGGSVDD1194 pKa = 5.17PIILDD1199 pKa = 3.71LNHH1202 pKa = 6.65NGFAFSDD1209 pKa = 4.03VSHH1212 pKa = 6.57GVQFDD1217 pKa = 3.73MNGDD1221 pKa = 3.63GTKK1224 pKa = 10.32EE1225 pKa = 3.48QLAWNTSHH1233 pKa = 7.83DD1234 pKa = 3.57GMLAVDD1240 pKa = 5.14LNHH1243 pKa = 7.37DD1244 pKa = 3.81GKK1246 pKa = 11.14INDD1249 pKa = 3.8GTEE1252 pKa = 3.75LFTPNFGGGNFASGVAALASLDD1274 pKa = 4.04SNHH1277 pKa = 7.34DD1278 pKa = 3.4GVIDD1282 pKa = 3.97HH1283 pKa = 7.23NDD1285 pKa = 3.14AAFSSLLIWKK1295 pKa = 8.42DD1296 pKa = 3.32ANANGISDD1304 pKa = 4.02AGEE1307 pKa = 4.11LSHH1310 pKa = 7.16LADD1313 pKa = 3.96NGVVSISTAAHH1324 pKa = 6.05PAVGEE1329 pKa = 3.93IDD1331 pKa = 3.77GQTVTGNGTFQMADD1345 pKa = 3.39GTSGNYY1351 pKa = 9.42VEE1353 pKa = 5.55VEE1355 pKa = 3.88LDD1357 pKa = 3.41TSLVAPTQPSVAIDD1371 pKa = 3.71GTSGADD1377 pKa = 3.11TFKK1380 pKa = 10.64IDD1382 pKa = 3.58NLNIKK1387 pKa = 10.09DD1388 pKa = 4.9LISDD1392 pKa = 3.4YY1393 pKa = 11.19HH1394 pKa = 6.96GAEE1397 pKa = 3.94GDD1399 pKa = 4.19KK1400 pKa = 10.46IDD1402 pKa = 5.61LSALFDD1408 pKa = 3.81RR1409 pKa = 11.84APAGNIADD1417 pKa = 3.75YY1418 pKa = 11.1VHH1420 pKa = 6.56YY1421 pKa = 10.79NSATSTVSVDD1431 pKa = 3.33TSGSGNPANFVDD1443 pKa = 4.89VAVLQNAPPVGTINILYY1460 pKa = 10.11DD1461 pKa = 3.73DD1462 pKa = 4.33TNHH1465 pKa = 6.38AQHH1468 pKa = 6.61TATII1472 pKa = 4.02
SS1 pKa = 5.77EE2 pKa = 4.3TTTGSITYY10 pKa = 7.68TAPDD14 pKa = 3.85GPATVTIGGLAVSAVGQTFAGSFGTLTITSIAAGSVGYY52 pKa = 10.45SYY54 pKa = 10.34TLTTNTSGDD63 pKa = 3.54TTHH66 pKa = 7.63DD67 pKa = 3.71DD68 pKa = 3.7FAVSVTDD75 pKa = 4.02ADD77 pKa = 4.39GDD79 pKa = 4.17HH80 pKa = 6.97SDD82 pKa = 3.4KK83 pKa = 11.0TLTINIVDD91 pKa = 4.45DD92 pKa = 4.18VPTAHH97 pKa = 7.6ADD99 pKa = 3.69TDD101 pKa = 4.26SVTAGSYY108 pKa = 10.69GPEE111 pKa = 4.14LGNVITAVGTTSSGADD127 pKa = 3.14TLGADD132 pKa = 3.94GAVVAGVAAGSGGNLDD148 pKa = 3.5NAATVGAVIHH158 pKa = 5.69GTYY161 pKa = 11.02GDD163 pKa = 4.11LTLQANGDD171 pKa = 3.67YY172 pKa = 10.71SYY174 pKa = 11.38ARR176 pKa = 11.84HH177 pKa = 6.44AGTPGGVNDD186 pKa = 3.59VFTYY190 pKa = 8.39TLKK193 pKa = 11.21DD194 pKa = 3.22GDD196 pKa = 4.49GDD198 pKa = 4.17LSHH201 pKa = 6.69TTLTISIGDD210 pKa = 3.77SGVTTTVPVAGTAGAQVSEE229 pKa = 4.85AGLPPHH235 pKa = 6.78GALPAGSGEE244 pKa = 4.44SADD247 pKa = 3.72GLPNNNSDD255 pKa = 3.44TSEE258 pKa = 4.17TTTGSITYY266 pKa = 7.68TAPDD270 pKa = 3.85GPATVTIGGLAVSAVGQTFAGSFGTLTITSIAAGSVGYY308 pKa = 10.45SYY310 pKa = 10.34TLTTNTSGDD319 pKa = 3.54TTHH322 pKa = 7.63DD323 pKa = 3.71DD324 pKa = 3.7FAVSVTDD331 pKa = 4.02ADD333 pKa = 4.39GDD335 pKa = 4.17HH336 pKa = 6.97SDD338 pKa = 3.4KK339 pKa = 11.0TLTINIVDD347 pKa = 4.45DD348 pKa = 4.18VPTAHH353 pKa = 7.26ADD355 pKa = 3.05SWAPAITAPTVLTGLLGNDD374 pKa = 3.07VFGADD379 pKa = 5.05GIATTTPGMVTTTNGAHH396 pKa = 5.83GTVTYY401 pKa = 11.11NNDD404 pKa = 2.79GTFTYY409 pKa = 9.37TPTGIYY415 pKa = 9.86VGSDD419 pKa = 2.42SFTYY423 pKa = 9.66TIKK426 pKa = 11.15DD427 pKa = 3.27GDD429 pKa = 4.42GDD431 pKa = 3.96TSTATVTVNVQTNTVPGGGGTASLTLNEE459 pKa = 4.55AALDD463 pKa = 4.08TTQDD467 pKa = 3.36AADD470 pKa = 3.91LHH472 pKa = 6.89AGAVTGTNPTSPGEE486 pKa = 4.21SAQATSGITFTTTGEE501 pKa = 4.49AITVAFANPTGDD513 pKa = 4.26PNWVAPAVSGLAAGYY528 pKa = 9.49SISWALSGGQLVGTLMQGVTNLGPAIYY555 pKa = 9.75LALSNTSAGANSSLTPVVTATLTDD579 pKa = 3.9QLQHH583 pKa = 6.07TAGSGNITINGLQVVATDD601 pKa = 3.35TSGDD605 pKa = 3.86HH606 pKa = 6.85VSGAVNLTVLDD617 pKa = 4.44DD618 pKa = 4.23APLPFVGDD626 pKa = 4.31SISLPDD632 pKa = 3.88TLHH635 pKa = 6.02TALTEE640 pKa = 4.23HH641 pKa = 7.02LNFAPGAGADD651 pKa = 3.65GVGNVVFNVTTGAAVQDD668 pKa = 3.51ASGANVYY675 pKa = 11.03LNGEE679 pKa = 3.88QLFYY683 pKa = 10.77QVSADD688 pKa = 3.02GHH690 pKa = 4.7TVEE693 pKa = 5.53GRR695 pKa = 11.84SSVANGNDD703 pKa = 3.24LGFTATLNPATDD715 pKa = 2.88TWSFVLNGTIFNGSQFTTAGATATGGNNQVNAFTALTSPDD755 pKa = 3.78TPNDD759 pKa = 3.69LLVTANGSNNVNTSTGNFGVGSGQSITSGEE789 pKa = 4.24TIRR792 pKa = 11.84FDD794 pKa = 3.93FVTNASTTGTIAGTNYY810 pKa = 5.62TTHH813 pKa = 7.09YY814 pKa = 9.0EE815 pKa = 4.2VSSFTQGITKK825 pKa = 10.64ANGTVDD831 pKa = 2.99ITIRR835 pKa = 11.84AVNADD840 pKa = 3.62DD841 pKa = 5.61DD842 pKa = 4.23KK843 pKa = 11.63TFVGDD848 pKa = 3.93GSGEE852 pKa = 3.9ATATGITVTFVNGSGGAAPTLTNNGDD878 pKa = 3.73GTFTLHH884 pKa = 7.45GLDD887 pKa = 3.73QGDD890 pKa = 4.07TFTITASSDD899 pKa = 3.17PFSAVEE905 pKa = 3.65ISGASSGTFKK915 pKa = 11.06LGAASFATANAVDD928 pKa = 4.52PFDD931 pKa = 4.29INIPVTGTDD940 pKa = 3.53GDD942 pKa = 4.08SDD944 pKa = 3.91TAAGSVTAHH953 pKa = 7.55LYY955 pKa = 7.03PTATTLEE962 pKa = 4.28GTNGVDD968 pKa = 3.24NLVATAVKK976 pKa = 9.59TILLGQDD983 pKa = 3.33GDD985 pKa = 4.05DD986 pKa = 4.13HH987 pKa = 6.3LTGINGHH994 pKa = 5.8DD995 pKa = 3.95TILSGGQGNDD1005 pKa = 3.04TLTGLDD1011 pKa = 4.25GKK1013 pKa = 9.72DD1014 pKa = 3.42TLIGGAGADD1023 pKa = 3.76TMTGGAGNDD1032 pKa = 3.53TFVIGAGEE1040 pKa = 4.26STPVIGGSGNAGTISGYY1057 pKa = 10.7DD1058 pKa = 3.2IITDD1062 pKa = 4.14FVAGTDD1068 pKa = 3.47KK1069 pKa = 10.68LTLPGSLVAATSGNVNGGDD1088 pKa = 3.52STLTIGGDD1096 pKa = 3.76TVQSHH1101 pKa = 5.73SVTNGIASFFGTDD1114 pKa = 3.35SFTAPLAVTSAASLAAVVQYY1134 pKa = 11.25LNATDD1139 pKa = 3.82IGNAGATLAFTATISGVNHH1158 pKa = 6.48TYY1160 pKa = 10.79VYY1162 pKa = 9.58TQATTNAGVGALVDD1176 pKa = 4.18LQGVTISNLNTLIGGSVDD1194 pKa = 5.17PIILDD1199 pKa = 3.71LNHH1202 pKa = 6.65NGFAFSDD1209 pKa = 4.03VSHH1212 pKa = 6.57GVQFDD1217 pKa = 3.73MNGDD1221 pKa = 3.63GTKK1224 pKa = 10.32EE1225 pKa = 3.48QLAWNTSHH1233 pKa = 7.83DD1234 pKa = 3.57GMLAVDD1240 pKa = 5.14LNHH1243 pKa = 7.37DD1244 pKa = 3.81GKK1246 pKa = 11.14INDD1249 pKa = 3.8GTEE1252 pKa = 3.75LFTPNFGGGNFASGVAALASLDD1274 pKa = 4.04SNHH1277 pKa = 7.34DD1278 pKa = 3.4GVIDD1282 pKa = 3.97HH1283 pKa = 7.23NDD1285 pKa = 3.14AAFSSLLIWKK1295 pKa = 8.42DD1296 pKa = 3.32ANANGISDD1304 pKa = 4.02AGEE1307 pKa = 4.11LSHH1310 pKa = 7.16LADD1313 pKa = 3.96NGVVSISTAAHH1324 pKa = 6.05PAVGEE1329 pKa = 3.93IDD1331 pKa = 3.77GQTVTGNGTFQMADD1345 pKa = 3.39GTSGNYY1351 pKa = 9.42VEE1353 pKa = 5.55VEE1355 pKa = 3.88LDD1357 pKa = 3.41TSLVAPTQPSVAIDD1371 pKa = 3.71GTSGADD1377 pKa = 3.11TFKK1380 pKa = 10.64IDD1382 pKa = 3.58NLNIKK1387 pKa = 10.09DD1388 pKa = 4.9LISDD1392 pKa = 3.4YY1393 pKa = 11.19HH1394 pKa = 6.96GAEE1397 pKa = 3.94GDD1399 pKa = 4.19KK1400 pKa = 10.46IDD1402 pKa = 5.61LSALFDD1408 pKa = 3.81RR1409 pKa = 11.84APAGNIADD1417 pKa = 3.75YY1418 pKa = 11.1VHH1420 pKa = 6.56YY1421 pKa = 10.79NSATSTVSVDD1431 pKa = 3.33TSGSGNPANFVDD1443 pKa = 4.89VAVLQNAPPVGTINILYY1460 pKa = 10.11DD1461 pKa = 3.73DD1462 pKa = 4.33TNHH1465 pKa = 6.38AQHH1468 pKa = 6.61TATII1472 pKa = 4.02
Molecular weight: 146.31 kDa
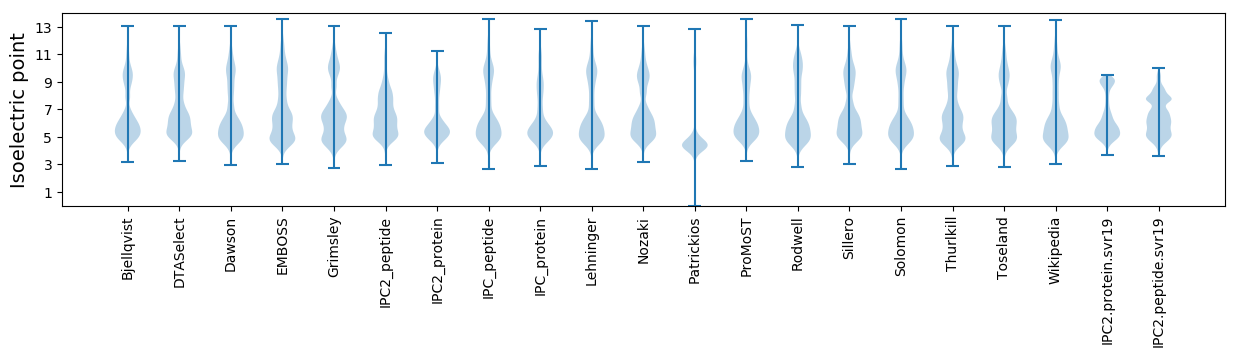
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A504DH39|A0A504DH39_9RHIZ CDP-alcohol phosphatidyltransferase family protein OS=Mesorhizobium sp. B2-3-3 OX=2589960 GN=FKO01_40620 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.51GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.51GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3950924 |
21 |
7542 |
319.5 |
34.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.978 ± 0.036 | 0.783 ± 0.007 |
5.911 ± 0.018 | 5.623 ± 0.019 |
3.253 ± 0.015 | 9.176 ± 0.019 |
2.162 ± 0.012 | 4.153 ± 0.022 |
2.879 ± 0.022 | 10.088 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.011 | 2.187 ± 0.012 |
5.598 ± 0.021 | 2.855 ± 0.012 |
7.446 ± 0.025 | 5.413 ± 0.016 |
5.784 ± 0.017 | 8.032 ± 0.021 |
1.416 ± 0.008 | 2.147 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
