
Bos taurus papillomavirus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Epsilonpapillomavirus; Epsilonpapillomavirus 1
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
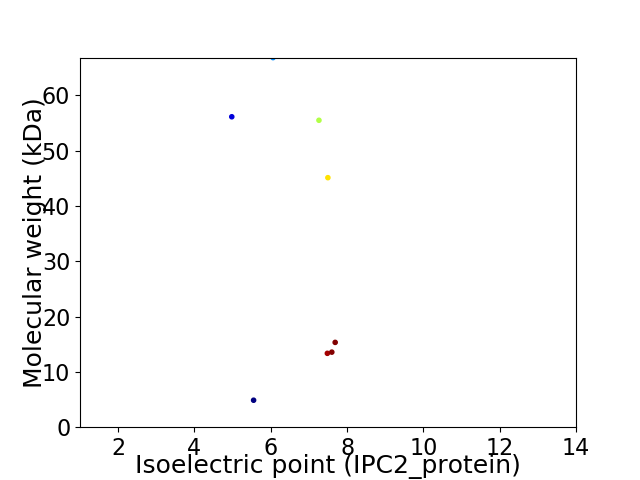
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6XA98|A6XA98_BPV5 Major capsid protein L1 OS=Bos taurus papillomavirus 8 OX=2758968 GN=L1 PE=3 SV=1
MM1 pKa = 7.47AVRR4 pKa = 11.84VRR6 pKa = 11.84RR7 pKa = 11.84VKK9 pKa = 10.41RR10 pKa = 11.84ANPYY14 pKa = 10.57DD15 pKa = 4.17LYY17 pKa = 9.98RR18 pKa = 11.84TCATGDD24 pKa = 3.64CPQDD28 pKa = 3.34VKK30 pKa = 11.59DD31 pKa = 3.92KK32 pKa = 11.21FEE34 pKa = 5.49HH35 pKa = 5.53NTLADD40 pKa = 5.16KK41 pKa = 10.46ILKK44 pKa = 8.89WGSTGIFLGGLGIGTGQGRR63 pKa = 11.84PGLGTYY69 pKa = 10.32SPLGRR74 pKa = 11.84GGGGGVSGRR83 pKa = 11.84IPTRR87 pKa = 11.84PSGSRR92 pKa = 11.84PLGRR96 pKa = 11.84PFNSGPIDD104 pKa = 3.66TIGAGGLRR112 pKa = 11.84PSVEE116 pKa = 4.13SSVNSADD123 pKa = 3.89VVALLPDD130 pKa = 3.87SPSVVTPDD138 pKa = 4.25SMPVDD143 pKa = 4.08PGVGGLDD150 pKa = 2.89IHH152 pKa = 8.29AEE154 pKa = 4.23VVEE157 pKa = 4.48EE158 pKa = 3.9PSLIFVEE165 pKa = 4.68PHH167 pKa = 4.84ATEE170 pKa = 4.44DD171 pKa = 4.2VAVLDD176 pKa = 4.33MNPAEE181 pKa = 4.84HH182 pKa = 6.91DD183 pKa = 3.54QSVHH187 pKa = 5.76LSSSTTHH194 pKa = 6.5HH195 pKa = 6.09NPSFQGHH202 pKa = 4.34VTLYY206 pKa = 9.87TDD208 pKa = 2.87IGEE211 pKa = 4.33TSEE214 pKa = 5.79VEE216 pKa = 4.11NLWIGGTSIGSNRR229 pKa = 11.84GEE231 pKa = 4.35EE232 pKa = 3.77IQMQLFSTPRR242 pKa = 11.84SSTPEE247 pKa = 3.5TEE249 pKa = 4.28AIGRR253 pKa = 11.84VRR255 pKa = 11.84GRR257 pKa = 11.84ANWFSRR263 pKa = 11.84RR264 pKa = 11.84YY265 pKa = 6.48YY266 pKa = 8.3TQVSVRR272 pKa = 11.84DD273 pKa = 3.97PKK275 pKa = 10.89FLEE278 pKa = 3.87QPRR281 pKa = 11.84EE282 pKa = 3.74YY283 pKa = 9.8FTYY286 pKa = 10.28GFEE289 pKa = 4.25NPAYY293 pKa = 9.83EE294 pKa = 4.71PDD296 pKa = 3.84PLEE299 pKa = 4.92DD300 pKa = 3.58SFDD303 pKa = 3.69VSEE306 pKa = 4.4VSPSVRR312 pKa = 11.84VEE314 pKa = 3.68PEE316 pKa = 3.63FRR318 pKa = 11.84DD319 pKa = 3.78LTHH322 pKa = 5.21VTAAKK327 pKa = 9.47TLTGNTGRR335 pKa = 11.84LGVSRR340 pKa = 11.84VGTKK344 pKa = 10.57LSIGTRR350 pKa = 11.84SGVRR354 pKa = 11.84IGGQVHH360 pKa = 5.95FRR362 pKa = 11.84YY363 pKa = 10.12SLSTIADD370 pKa = 3.69EE371 pKa = 5.18EE372 pKa = 4.39GLEE375 pKa = 3.95LLPRR379 pKa = 11.84SAAEE383 pKa = 4.26GSAGPLQHH391 pKa = 6.8SAEE394 pKa = 4.31TVLTDD399 pKa = 3.06GHH401 pKa = 6.29DD402 pKa = 3.5AYY404 pKa = 11.7VEE406 pKa = 3.98VDD408 pKa = 3.42HH409 pKa = 7.01NSTGSAYY416 pKa = 10.78SEE418 pKa = 3.86IDD420 pKa = 3.66LLDD423 pKa = 3.99EE424 pKa = 5.91DD425 pKa = 4.92EE426 pKa = 4.47VTPHH430 pKa = 5.98GVLVFHH436 pKa = 7.25DD437 pKa = 5.01RR438 pKa = 11.84PAEE441 pKa = 3.93TDD443 pKa = 3.62VVPITDD449 pKa = 2.96VSYY452 pKa = 10.96AIKK455 pKa = 10.02PLSTIPGDD463 pKa = 3.95DD464 pKa = 4.69LLPSSTITMQNGPVHH479 pKa = 7.24VDD481 pKa = 3.19GQDD484 pKa = 3.33NIVPDD489 pKa = 4.42IIIIDD494 pKa = 3.99FSVEE498 pKa = 3.28GTYY501 pKa = 10.68FLNTYY506 pKa = 8.86AHH508 pKa = 7.08PSLHH512 pKa = 5.76KK513 pKa = 9.88RR514 pKa = 11.84KK515 pKa = 9.67KK516 pKa = 9.9RR517 pKa = 11.84RR518 pKa = 11.84LSS520 pKa = 3.29
MM1 pKa = 7.47AVRR4 pKa = 11.84VRR6 pKa = 11.84RR7 pKa = 11.84VKK9 pKa = 10.41RR10 pKa = 11.84ANPYY14 pKa = 10.57DD15 pKa = 4.17LYY17 pKa = 9.98RR18 pKa = 11.84TCATGDD24 pKa = 3.64CPQDD28 pKa = 3.34VKK30 pKa = 11.59DD31 pKa = 3.92KK32 pKa = 11.21FEE34 pKa = 5.49HH35 pKa = 5.53NTLADD40 pKa = 5.16KK41 pKa = 10.46ILKK44 pKa = 8.89WGSTGIFLGGLGIGTGQGRR63 pKa = 11.84PGLGTYY69 pKa = 10.32SPLGRR74 pKa = 11.84GGGGGVSGRR83 pKa = 11.84IPTRR87 pKa = 11.84PSGSRR92 pKa = 11.84PLGRR96 pKa = 11.84PFNSGPIDD104 pKa = 3.66TIGAGGLRR112 pKa = 11.84PSVEE116 pKa = 4.13SSVNSADD123 pKa = 3.89VVALLPDD130 pKa = 3.87SPSVVTPDD138 pKa = 4.25SMPVDD143 pKa = 4.08PGVGGLDD150 pKa = 2.89IHH152 pKa = 8.29AEE154 pKa = 4.23VVEE157 pKa = 4.48EE158 pKa = 3.9PSLIFVEE165 pKa = 4.68PHH167 pKa = 4.84ATEE170 pKa = 4.44DD171 pKa = 4.2VAVLDD176 pKa = 4.33MNPAEE181 pKa = 4.84HH182 pKa = 6.91DD183 pKa = 3.54QSVHH187 pKa = 5.76LSSSTTHH194 pKa = 6.5HH195 pKa = 6.09NPSFQGHH202 pKa = 4.34VTLYY206 pKa = 9.87TDD208 pKa = 2.87IGEE211 pKa = 4.33TSEE214 pKa = 5.79VEE216 pKa = 4.11NLWIGGTSIGSNRR229 pKa = 11.84GEE231 pKa = 4.35EE232 pKa = 3.77IQMQLFSTPRR242 pKa = 11.84SSTPEE247 pKa = 3.5TEE249 pKa = 4.28AIGRR253 pKa = 11.84VRR255 pKa = 11.84GRR257 pKa = 11.84ANWFSRR263 pKa = 11.84RR264 pKa = 11.84YY265 pKa = 6.48YY266 pKa = 8.3TQVSVRR272 pKa = 11.84DD273 pKa = 3.97PKK275 pKa = 10.89FLEE278 pKa = 3.87QPRR281 pKa = 11.84EE282 pKa = 3.74YY283 pKa = 9.8FTYY286 pKa = 10.28GFEE289 pKa = 4.25NPAYY293 pKa = 9.83EE294 pKa = 4.71PDD296 pKa = 3.84PLEE299 pKa = 4.92DD300 pKa = 3.58SFDD303 pKa = 3.69VSEE306 pKa = 4.4VSPSVRR312 pKa = 11.84VEE314 pKa = 3.68PEE316 pKa = 3.63FRR318 pKa = 11.84DD319 pKa = 3.78LTHH322 pKa = 5.21VTAAKK327 pKa = 9.47TLTGNTGRR335 pKa = 11.84LGVSRR340 pKa = 11.84VGTKK344 pKa = 10.57LSIGTRR350 pKa = 11.84SGVRR354 pKa = 11.84IGGQVHH360 pKa = 5.95FRR362 pKa = 11.84YY363 pKa = 10.12SLSTIADD370 pKa = 3.69EE371 pKa = 5.18EE372 pKa = 4.39GLEE375 pKa = 3.95LLPRR379 pKa = 11.84SAAEE383 pKa = 4.26GSAGPLQHH391 pKa = 6.8SAEE394 pKa = 4.31TVLTDD399 pKa = 3.06GHH401 pKa = 6.29DD402 pKa = 3.5AYY404 pKa = 11.7VEE406 pKa = 3.98VDD408 pKa = 3.42HH409 pKa = 7.01NSTGSAYY416 pKa = 10.78SEE418 pKa = 3.86IDD420 pKa = 3.66LLDD423 pKa = 3.99EE424 pKa = 5.91DD425 pKa = 4.92EE426 pKa = 4.47VTPHH430 pKa = 5.98GVLVFHH436 pKa = 7.25DD437 pKa = 5.01RR438 pKa = 11.84PAEE441 pKa = 3.93TDD443 pKa = 3.62VVPITDD449 pKa = 2.96VSYY452 pKa = 10.96AIKK455 pKa = 10.02PLSTIPGDD463 pKa = 3.95DD464 pKa = 4.69LLPSSTITMQNGPVHH479 pKa = 7.24VDD481 pKa = 3.19GQDD484 pKa = 3.33NIVPDD489 pKa = 4.42IIIIDD494 pKa = 3.99FSVEE498 pKa = 3.28GTYY501 pKa = 10.68FLNTYY506 pKa = 8.86AHH508 pKa = 7.08PSLHH512 pKa = 5.76KK513 pKa = 9.88RR514 pKa = 11.84KK515 pKa = 9.67KK516 pKa = 9.9RR517 pKa = 11.84RR518 pKa = 11.84LSS520 pKa = 3.29
Molecular weight: 56.13 kDa
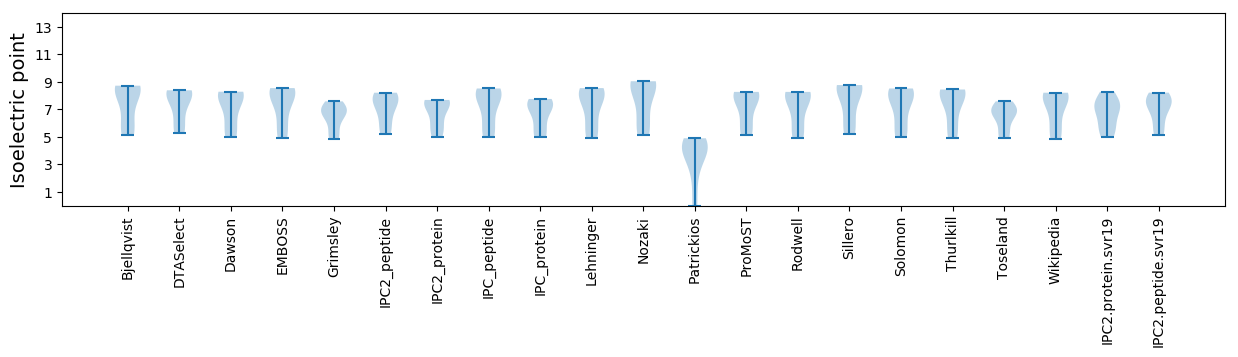
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6XA92|A6XA92_BPV5 E7 OS=Bos taurus papillomavirus 8 OX=2758968 PE=4 SV=1
MM1 pKa = 7.72PCFIPSYY8 pKa = 10.83NFDD11 pKa = 3.81GLDD14 pKa = 3.52CLVCKK19 pKa = 10.46RR20 pKa = 11.84PLGSYY25 pKa = 10.13DD26 pKa = 3.55ALKK29 pKa = 10.94CKK31 pKa = 8.39TNKK34 pKa = 9.05FRR36 pKa = 11.84RR37 pKa = 11.84VHH39 pKa = 6.68RR40 pKa = 11.84DD41 pKa = 2.6GHH43 pKa = 6.24VYY45 pKa = 10.71GMCCGCTEE53 pKa = 4.65LLLKK57 pKa = 10.13IEE59 pKa = 4.84RR60 pKa = 11.84EE61 pKa = 4.17DD62 pKa = 3.93NPWSLVCPHH71 pKa = 7.07DD72 pKa = 3.84FKK74 pKa = 11.41KK75 pKa = 11.1VFGRR79 pKa = 11.84QVGDD83 pKa = 3.95CIVRR87 pKa = 11.84CYY89 pKa = 11.22YY90 pKa = 10.29CGCPLSDD97 pKa = 3.93SEE99 pKa = 4.8KK100 pKa = 10.64DD101 pKa = 3.2RR102 pKa = 11.84HH103 pKa = 5.69ALASEE108 pKa = 5.29GYY110 pKa = 10.03LFLRR114 pKa = 11.84GKK116 pKa = 9.72PRR118 pKa = 11.84GRR120 pKa = 11.84CYY122 pKa = 11.05ACASDD127 pKa = 3.77GRR129 pKa = 11.84RR130 pKa = 11.84PCLYY134 pKa = 10.67
MM1 pKa = 7.72PCFIPSYY8 pKa = 10.83NFDD11 pKa = 3.81GLDD14 pKa = 3.52CLVCKK19 pKa = 10.46RR20 pKa = 11.84PLGSYY25 pKa = 10.13DD26 pKa = 3.55ALKK29 pKa = 10.94CKK31 pKa = 8.39TNKK34 pKa = 9.05FRR36 pKa = 11.84RR37 pKa = 11.84VHH39 pKa = 6.68RR40 pKa = 11.84DD41 pKa = 2.6GHH43 pKa = 6.24VYY45 pKa = 10.71GMCCGCTEE53 pKa = 4.65LLLKK57 pKa = 10.13IEE59 pKa = 4.84RR60 pKa = 11.84EE61 pKa = 4.17DD62 pKa = 3.93NPWSLVCPHH71 pKa = 7.07DD72 pKa = 3.84FKK74 pKa = 11.41KK75 pKa = 11.1VFGRR79 pKa = 11.84QVGDD83 pKa = 3.95CIVRR87 pKa = 11.84CYY89 pKa = 11.22YY90 pKa = 10.29CGCPLSDD97 pKa = 3.93SEE99 pKa = 4.8KK100 pKa = 10.64DD101 pKa = 3.2RR102 pKa = 11.84HH103 pKa = 5.69ALASEE108 pKa = 5.29GYY110 pKa = 10.03LFLRR114 pKa = 11.84GKK116 pKa = 9.72PRR118 pKa = 11.84GRR120 pKa = 11.84CYY122 pKa = 11.05ACASDD127 pKa = 3.77GRR129 pKa = 11.84RR130 pKa = 11.84PCLYY134 pKa = 10.67
Molecular weight: 15.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2439 |
40 |
593 |
304.9 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.191 ± 0.588 | 3.034 ± 1.195 |
5.74 ± 0.409 | 5.453 ± 0.395 |
4.223 ± 0.417 | 7.749 ± 0.905 |
2.788 ± 0.523 | 3.772 ± 0.384 |
4.592 ± 0.685 | 8.979 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.517 ± 0.238 | 3.895 ± 0.743 |
6.765 ± 0.943 | 4.059 ± 0.693 |
6.191 ± 0.754 | 7.626 ± 0.687 |
6.56 ± 0.558 | 6.56 ± 0.637 |
1.312 ± 0.238 | 2.993 ± 0.415 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
