
Sewage-associated circular DNA virus-15
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
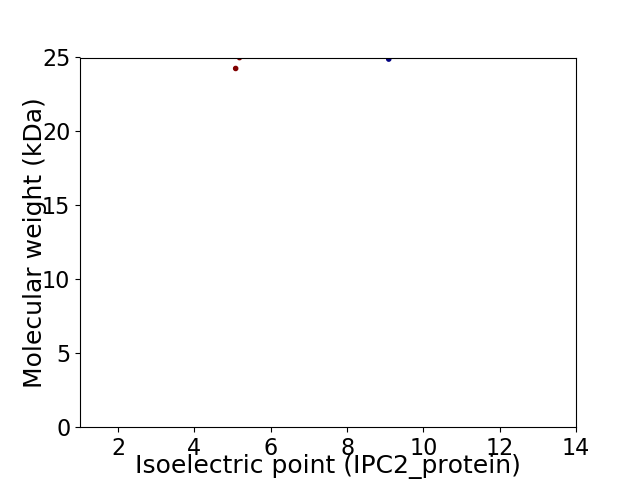
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UGX1|A0A0B4UGX1_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-15 OX=1592082 PE=4 SV=1
MM1 pKa = 7.91GDD3 pKa = 3.16TSQRR7 pKa = 11.84ATCWSITINNPLPSDD22 pKa = 3.51IPTAEE27 pKa = 4.35ALPAKK32 pKa = 9.35WVLQGQLEE40 pKa = 4.23EE41 pKa = 4.47GKK43 pKa = 10.15EE44 pKa = 4.37GTVHH48 pKa = 4.86YY49 pKa = 10.34QGMLTTPQCRR59 pKa = 11.84FSQVKK64 pKa = 9.65KK65 pKa = 9.63VLPRR69 pKa = 11.84AHH71 pKa = 6.92IEE73 pKa = 3.89VAKK76 pKa = 10.89NRR78 pKa = 11.84TALEE82 pKa = 4.32KK83 pKa = 10.63YY84 pKa = 7.4VHH86 pKa = 6.76KK87 pKa = 10.55EE88 pKa = 3.4DD89 pKa = 3.58TRR91 pKa = 11.84LVEE94 pKa = 4.23VPTVSSSLPTLFDD107 pKa = 3.36YY108 pKa = 11.18QFTVAGRR115 pKa = 11.84WVSTEE120 pKa = 3.31FDD122 pKa = 3.47KK123 pKa = 11.87YY124 pKa = 11.02IDD126 pKa = 4.03DD127 pKa = 5.65FKK129 pKa = 11.64DD130 pKa = 3.27KK131 pKa = 11.06DD132 pKa = 3.54IGDD135 pKa = 3.75VALLYY140 pKa = 10.65VDD142 pKa = 4.57SLVEE146 pKa = 3.76KK147 pKa = 10.61DD148 pKa = 4.24IINGMRR154 pKa = 11.84GIEE157 pKa = 4.41FIAINPMWRR166 pKa = 11.84SSWKK170 pKa = 10.1RR171 pKa = 11.84FWRR174 pKa = 11.84GIILRR179 pKa = 11.84HH180 pKa = 6.09RR181 pKa = 11.84SIEE184 pKa = 4.11DD185 pKa = 3.09AQARR189 pKa = 11.84QEE191 pKa = 4.82DD192 pKa = 4.16IPSQEE197 pKa = 4.09EE198 pKa = 4.49CTASPCSEE206 pKa = 4.03AQDD209 pKa = 4.49GEE211 pKa = 4.72CEE213 pKa = 3.89
MM1 pKa = 7.91GDD3 pKa = 3.16TSQRR7 pKa = 11.84ATCWSITINNPLPSDD22 pKa = 3.51IPTAEE27 pKa = 4.35ALPAKK32 pKa = 9.35WVLQGQLEE40 pKa = 4.23EE41 pKa = 4.47GKK43 pKa = 10.15EE44 pKa = 4.37GTVHH48 pKa = 4.86YY49 pKa = 10.34QGMLTTPQCRR59 pKa = 11.84FSQVKK64 pKa = 9.65KK65 pKa = 9.63VLPRR69 pKa = 11.84AHH71 pKa = 6.92IEE73 pKa = 3.89VAKK76 pKa = 10.89NRR78 pKa = 11.84TALEE82 pKa = 4.32KK83 pKa = 10.63YY84 pKa = 7.4VHH86 pKa = 6.76KK87 pKa = 10.55EE88 pKa = 3.4DD89 pKa = 3.58TRR91 pKa = 11.84LVEE94 pKa = 4.23VPTVSSSLPTLFDD107 pKa = 3.36YY108 pKa = 11.18QFTVAGRR115 pKa = 11.84WVSTEE120 pKa = 3.31FDD122 pKa = 3.47KK123 pKa = 11.87YY124 pKa = 11.02IDD126 pKa = 4.03DD127 pKa = 5.65FKK129 pKa = 11.64DD130 pKa = 3.27KK131 pKa = 11.06DD132 pKa = 3.54IGDD135 pKa = 3.75VALLYY140 pKa = 10.65VDD142 pKa = 4.57SLVEE146 pKa = 3.76KK147 pKa = 10.61DD148 pKa = 4.24IINGMRR154 pKa = 11.84GIEE157 pKa = 4.41FIAINPMWRR166 pKa = 11.84SSWKK170 pKa = 10.1RR171 pKa = 11.84FWRR174 pKa = 11.84GIILRR179 pKa = 11.84HH180 pKa = 6.09RR181 pKa = 11.84SIEE184 pKa = 4.11DD185 pKa = 3.09AQARR189 pKa = 11.84QEE191 pKa = 4.82DD192 pKa = 4.16IPSQEE197 pKa = 4.09EE198 pKa = 4.49CTASPCSEE206 pKa = 4.03AQDD209 pKa = 4.49GEE211 pKa = 4.72CEE213 pKa = 3.89
Molecular weight: 24.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UGX1|A0A0B4UGX1_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-15 OX=1592082 PE=4 SV=1
MM1 pKa = 7.69PKK3 pKa = 10.0LVKK6 pKa = 9.19KK7 pKa = 8.82TYY9 pKa = 9.21PRR11 pKa = 11.84KK12 pKa = 9.17KK13 pKa = 7.9NVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84VAKK22 pKa = 10.25HH23 pKa = 4.2RR24 pKa = 11.84TVNVNRR30 pKa = 11.84ALQPFAQRR38 pKa = 11.84YY39 pKa = 4.58ITKK42 pKa = 9.03MKK44 pKa = 10.47YY45 pKa = 10.0SEE47 pKa = 5.49AYY49 pKa = 8.3TLNAGNIWSQIMNLNSIFDD68 pKa = 4.3PNRR71 pKa = 11.84TGIGHH76 pKa = 5.8QPYY79 pKa = 10.47GHH81 pKa = 7.17DD82 pKa = 3.84TLQQIYY88 pKa = 10.17GRR90 pKa = 11.84YY91 pKa = 8.84RR92 pKa = 11.84VISCKK97 pKa = 10.58YY98 pKa = 10.0VVNCYY103 pKa = 10.28NATTPIRR110 pKa = 11.84FGCLPANEE118 pKa = 4.69IPPVSTVSEE127 pKa = 4.18LCEE130 pKa = 3.87NPRR133 pKa = 11.84SQFRR137 pKa = 11.84IQFPGGSTQTIAGVVSIPSLVGRR160 pKa = 11.84NKK162 pKa = 9.78SQYY165 pKa = 8.71MADD168 pKa = 4.44DD169 pKa = 4.59RR170 pKa = 11.84FQAPFGSSPAEE181 pKa = 3.7LALLYY186 pKa = 9.57ITGQSMADD194 pKa = 3.42SNVDD198 pKa = 2.77INLNVTLEE206 pKa = 3.96YY207 pKa = 10.59LVEE210 pKa = 4.41CFDD213 pKa = 5.32VIPLEE218 pKa = 4.24QSS220 pKa = 2.86
MM1 pKa = 7.69PKK3 pKa = 10.0LVKK6 pKa = 9.19KK7 pKa = 8.82TYY9 pKa = 9.21PRR11 pKa = 11.84KK12 pKa = 9.17KK13 pKa = 7.9NVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84VAKK22 pKa = 10.25HH23 pKa = 4.2RR24 pKa = 11.84TVNVNRR30 pKa = 11.84ALQPFAQRR38 pKa = 11.84YY39 pKa = 4.58ITKK42 pKa = 9.03MKK44 pKa = 10.47YY45 pKa = 10.0SEE47 pKa = 5.49AYY49 pKa = 8.3TLNAGNIWSQIMNLNSIFDD68 pKa = 4.3PNRR71 pKa = 11.84TGIGHH76 pKa = 5.8QPYY79 pKa = 10.47GHH81 pKa = 7.17DD82 pKa = 3.84TLQQIYY88 pKa = 10.17GRR90 pKa = 11.84YY91 pKa = 8.84RR92 pKa = 11.84VISCKK97 pKa = 10.58YY98 pKa = 10.0VVNCYY103 pKa = 10.28NATTPIRR110 pKa = 11.84FGCLPANEE118 pKa = 4.69IPPVSTVSEE127 pKa = 4.18LCEE130 pKa = 3.87NPRR133 pKa = 11.84SQFRR137 pKa = 11.84IQFPGGSTQTIAGVVSIPSLVGRR160 pKa = 11.84NKK162 pKa = 9.78SQYY165 pKa = 8.71MADD168 pKa = 4.44DD169 pKa = 4.59RR170 pKa = 11.84FQAPFGSSPAEE181 pKa = 3.7LALLYY186 pKa = 9.57ITGQSMADD194 pKa = 3.42SNVDD198 pKa = 2.77INLNVTLEE206 pKa = 3.96YY207 pKa = 10.59LVEE210 pKa = 4.41CFDD213 pKa = 5.32VIPLEE218 pKa = 4.24QSS220 pKa = 2.86
Molecular weight: 24.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
433 |
213 |
220 |
216.5 |
24.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.236 ± 0.236 | 2.309 ± 0.027 |
5.081 ± 1.37 | 6.005 ± 1.709 |
3.464 ± 0.124 | 5.312 ± 0.103 |
1.617 ± 0.183 | 6.928 ± 0.08 |
5.081 ± 0.386 | 6.697 ± 0.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.079 ± 0.14 | 4.85 ± 1.748 |
6.005 ± 0.587 | 5.543 ± 0.264 |
6.697 ± 0.415 | 7.159 ± 0.082 |
6.236 ± 0.236 | 7.159 ± 0.41 |
1.617 ± 0.839 | 3.926 ± 1.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
