
Zeaxanthinibacter enoshimensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Zeaxanthinibacter
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
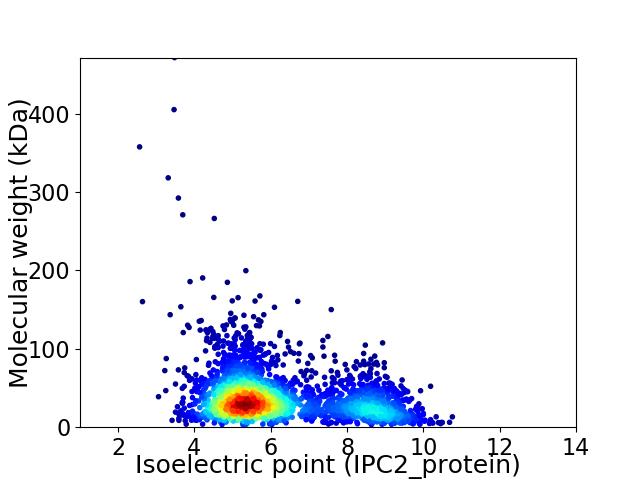
Virtual 2D-PAGE plot for 2926 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6TPB2|A0A4R6TPB2_9FLAO TPR_REGION domain-containing protein OS=Zeaxanthinibacter enoshimensis OX=392009 GN=CLV82_1906 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.76NKK4 pKa = 8.5ITRR7 pKa = 11.84SNLTPFILNTIPANFTKK24 pKa = 10.34ILAFVIIVFLSNNLLQAQLPQQFQKK49 pKa = 11.05VEE51 pKa = 4.9LITEE55 pKa = 4.01MTNAVNFEE63 pKa = 4.08FAPDD67 pKa = 3.38GRR69 pKa = 11.84IFIVDD74 pKa = 4.0RR75 pKa = 11.84YY76 pKa = 10.97GEE78 pKa = 4.0LLIYY82 pKa = 10.42NPNTQVTVSAGTLEE96 pKa = 4.15VFKK99 pKa = 11.03DD100 pKa = 4.1FEE102 pKa = 6.37DD103 pKa = 4.36GLLGVAFDD111 pKa = 5.62PGFLTNNYY119 pKa = 9.32IYY121 pKa = 10.95LHH123 pKa = 5.72YY124 pKa = 10.87SPLNVSVNRR133 pKa = 11.84VSRR136 pKa = 11.84FTMNGNTLDD145 pKa = 3.91LASEE149 pKa = 4.41VTLMQWPTQRR159 pKa = 11.84NGCCHH164 pKa = 6.72SGGDD168 pKa = 3.99MGFDD172 pKa = 3.57SQGNLYY178 pKa = 10.02IATGDD183 pKa = 3.71NANHH187 pKa = 6.63SDD189 pKa = 3.54YY190 pKa = 11.64AAISEE195 pKa = 4.57TNSVNSAEE203 pKa = 4.35KK204 pKa = 10.66SSSNTNDD211 pKa = 2.95LRR213 pKa = 11.84GKK215 pKa = 9.56ILRR218 pKa = 11.84VRR220 pKa = 11.84PQPDD224 pKa = 3.09GSYY227 pKa = 9.74TIPGGNLFPNGAGGLPEE244 pKa = 4.64IYY246 pKa = 11.06VMGARR251 pKa = 11.84NPYY254 pKa = 10.65RR255 pKa = 11.84MFIDD259 pKa = 4.44KK260 pKa = 10.76EE261 pKa = 4.05NTDD264 pKa = 3.66WIFWGEE270 pKa = 4.14VGPDD274 pKa = 3.21ANVASSAGPEE284 pKa = 4.19GKK286 pKa = 10.12DD287 pKa = 3.53EE288 pKa = 4.52INLTKK293 pKa = 10.22TAGNYY298 pKa = 6.68GWPYY302 pKa = 10.18FSGNNEE308 pKa = 4.35PYY310 pKa = 10.64LIDD313 pKa = 3.6YY314 pKa = 7.72TNPQYY319 pKa = 11.6YY320 pKa = 10.11NDD322 pKa = 4.69PNTPQNVSTWNTGATNLPPARR343 pKa = 11.84PSWLDD348 pKa = 3.53FFHH351 pKa = 7.06KK352 pKa = 10.4SYY354 pKa = 10.27MAGPRR359 pKa = 11.84YY360 pKa = 9.92YY361 pKa = 10.77YY362 pKa = 10.93NPALTDD368 pKa = 3.49QQRR371 pKa = 11.84LPIEE375 pKa = 3.97YY376 pKa = 10.25DD377 pKa = 3.28EE378 pKa = 4.53VFFYY382 pKa = 11.15FDD384 pKa = 4.12FNSSAVWGVKK394 pKa = 8.46MDD396 pKa = 3.57EE397 pKa = 4.05QGIIQSNQRR406 pKa = 11.84LNTAFPANNNLGFIDD421 pKa = 4.13MKK423 pKa = 10.49IGPDD427 pKa = 2.92GHH429 pKa = 8.55LYY431 pKa = 9.7ILEE434 pKa = 4.35YY435 pKa = 10.91GAGCCPNNTGTGKK448 pKa = 10.27LVRR451 pKa = 11.84MDD453 pKa = 3.76YY454 pKa = 10.75IGIVSNSSPVVEE466 pKa = 5.51ISADD470 pKa = 3.63PDD472 pKa = 3.65NGSLPLTVNFSSEE485 pKa = 4.1GTFDD489 pKa = 4.23PDD491 pKa = 3.62GDD493 pKa = 4.17SLTYY497 pKa = 10.24EE498 pKa = 4.17WDD500 pKa = 3.49FEE502 pKa = 4.54TDD504 pKa = 3.21GTVDD508 pKa = 3.37STEE511 pKa = 3.95EE512 pKa = 3.96NPTFVYY518 pKa = 10.75SLAGTYY524 pKa = 9.47NVQLRR529 pKa = 11.84VDD531 pKa = 4.19DD532 pKa = 4.63GNGGVSTKK540 pKa = 10.87NLTIYY545 pKa = 10.44AGNNAPTFTFTSPIDD560 pKa = 3.61GGFMNWGDD568 pKa = 5.75DD569 pKa = 3.24IDD571 pKa = 6.35LDD573 pKa = 4.34LLVEE577 pKa = 4.23DD578 pKa = 4.94VEE580 pKa = 5.69DD581 pKa = 4.47GSTANGGINCLDD593 pKa = 3.81VNVVPSLGHH602 pKa = 6.87LNHH605 pKa = 6.21FHH607 pKa = 7.96DD608 pKa = 6.17DD609 pKa = 3.72MGLDD613 pKa = 4.1GCPKK617 pKa = 9.75TLNLDD622 pKa = 3.9PGNHH626 pKa = 6.53NIYY629 pKa = 11.09GEE631 pKa = 3.81MDD633 pKa = 3.01IFYY636 pKa = 10.85VLGANYY642 pKa = 9.0TDD644 pKa = 3.48QGGLQAFDD652 pKa = 3.33QVQLHH657 pKa = 5.88PKK659 pKa = 9.18RR660 pKa = 11.84RR661 pKa = 11.84EE662 pKa = 3.63AEE664 pKa = 4.19FYY666 pKa = 10.51DD667 pKa = 3.91VKK669 pKa = 11.39SDD671 pKa = 3.2ITVVGNSDD679 pKa = 2.91PWGGGSEE686 pKa = 4.72AIRR689 pKa = 11.84VNHH692 pKa = 6.95DD693 pKa = 3.19GYY695 pKa = 10.79IVFEE699 pKa = 4.13GRR701 pKa = 11.84NLLNISGVKK710 pKa = 9.83YY711 pKa = 10.01RR712 pKa = 11.84VASQTAGGSIEE723 pKa = 4.54LRR725 pKa = 11.84VGSPTGALLATSPVPSTGSLNNWVDD750 pKa = 3.56VEE752 pKa = 4.51SAISDD757 pKa = 3.58PGGKK761 pKa = 9.4NDD763 pKa = 4.6LYY765 pKa = 11.18FVFKK769 pKa = 10.52NASAGVDD776 pKa = 3.21IFDD779 pKa = 4.33LNYY782 pKa = 10.09IEE784 pKa = 6.26FIGQGVSTDD793 pKa = 3.31NTPPVVNNVSSSEE806 pKa = 3.93PTEE809 pKa = 3.83VRR811 pKa = 11.84VEE813 pKa = 3.87FSEE816 pKa = 4.36YY817 pKa = 11.06VNMTVAEE824 pKa = 4.6TITNYY829 pKa = 10.87SLDD832 pKa = 3.54NGEE835 pKa = 4.55SVISASLQEE844 pKa = 4.29DD845 pKa = 3.48QRR847 pKa = 11.84TVILTTSQLAPGITYY862 pKa = 9.12TLDD865 pKa = 3.1INNVTNEE872 pKa = 3.74AGLPVASGSYY882 pKa = 8.75TFNFEE887 pKa = 4.2VEE889 pKa = 4.61CNNTPFPDD897 pKa = 3.09QWQEE901 pKa = 3.84EE902 pKa = 4.83IIGSEE907 pKa = 3.87LPYY910 pKa = 10.31RR911 pKa = 11.84SVYY914 pKa = 10.16IYY916 pKa = 10.88ADD918 pKa = 3.55HH919 pKa = 7.58DD920 pKa = 4.38LDD922 pKa = 6.01GDD924 pKa = 4.03GLKK927 pKa = 10.82DD928 pKa = 3.25IVTGGWWYY936 pKa = 10.82KK937 pKa = 11.09NPGTVTGNWVRR948 pKa = 11.84STIGSPFNNVAYY960 pKa = 10.64VYY962 pKa = 10.87DD963 pKa = 4.06FDD965 pKa = 6.69GDD967 pKa = 3.67GDD969 pKa = 4.27FDD971 pKa = 5.3LLGTQGAYY979 pKa = 9.92EE980 pKa = 4.42GSDD983 pKa = 3.36MAWAQNDD990 pKa = 3.6GSGNFTVYY998 pKa = 10.8TNIDD1002 pKa = 3.7SGQTTYY1008 pKa = 11.04VEE1010 pKa = 4.3PFLAGIAGGVFDD1022 pKa = 4.0NTGDD1026 pKa = 3.59FKK1028 pKa = 11.5LAINWNGAEE1037 pKa = 4.25SSGSPVQLLTIPADD1051 pKa = 3.5PVNTQWTFEE1060 pKa = 4.34NISPDD1065 pKa = 3.56SLGEE1069 pKa = 3.93ALSAGDD1075 pKa = 4.37IDD1077 pKa = 6.36DD1078 pKa = 6.36DD1079 pKa = 5.01GDD1081 pKa = 4.15LDD1083 pKa = 5.28LFQGSNWLRR1092 pKa = 11.84NEE1094 pKa = 4.37GNGTWTTFSTGITYY1108 pKa = 7.76PTTMDD1113 pKa = 3.45RR1114 pKa = 11.84NMLADD1119 pKa = 4.43FDD1121 pKa = 5.32RR1122 pKa = 11.84DD1123 pKa = 3.33GDD1125 pKa = 3.8LDD1127 pKa = 3.94AVVSQLGSNQEE1138 pKa = 4.1IAWFEE1143 pKa = 3.97APEE1146 pKa = 5.18DD1147 pKa = 3.91PTQPWTKK1154 pKa = 9.24NTLDD1158 pKa = 3.45TDD1160 pKa = 3.42IDD1162 pKa = 3.91GGLSVDD1168 pKa = 4.69VIDD1171 pKa = 5.94FDD1173 pKa = 5.11FDD1175 pKa = 3.6GDD1177 pKa = 3.56ADD1179 pKa = 3.73IVVGEE1184 pKa = 4.16WQSAYY1189 pKa = 10.94RR1190 pKa = 11.84LFAFEE1195 pKa = 5.71NDD1197 pKa = 3.48LCNTGTWIKK1206 pKa = 9.26HH1207 pKa = 5.38TIHH1210 pKa = 7.24PGTPTFDD1217 pKa = 3.38HH1218 pKa = 7.21HH1219 pKa = 7.72DD1220 pKa = 3.69GAQVVDD1226 pKa = 3.55IDD1228 pKa = 4.28EE1229 pKa = 5.94DD1230 pKa = 4.33GDD1232 pKa = 4.22LDD1234 pKa = 3.87IVSIGWNNITPRR1246 pKa = 11.84IFEE1249 pKa = 4.31NTTGSAPVNSPPMVLNPGDD1268 pKa = 3.43QLFNEE1273 pKa = 4.71GAIVNLQIQGSDD1285 pKa = 3.35PDD1287 pKa = 4.02FSDD1290 pKa = 3.17VLTYY1294 pKa = 9.03TAQGLPADD1302 pKa = 4.77LSIDD1306 pKa = 3.89PDD1308 pKa = 3.64TGIIGGNLSAAQGSYY1323 pKa = 10.15PVTVRR1328 pKa = 11.84VTDD1331 pKa = 3.32NGGLFDD1337 pKa = 4.17EE1338 pKa = 4.88TTFTIFVGAFNSIIRR1353 pKa = 11.84INAGGPLVTVNNEE1366 pKa = 3.24DD1367 pKa = 3.89WIADD1371 pKa = 3.41QYY1373 pKa = 11.56FNGGATFSTANAINGTTDD1391 pKa = 4.22DD1392 pKa = 4.04ILYY1395 pKa = 8.01QTEE1398 pKa = 4.36RR1399 pKa = 11.84NSDD1402 pKa = 4.05DD1403 pKa = 3.26PTLTYY1408 pKa = 10.02QIPVPEE1414 pKa = 4.19NGEE1417 pKa = 3.98YY1418 pKa = 10.63GVRR1421 pKa = 11.84LLFAEE1426 pKa = 4.68IFHH1429 pKa = 6.7NSTGSRR1435 pKa = 11.84LFDD1438 pKa = 3.36VDD1440 pKa = 3.35IEE1442 pKa = 4.52GGQGQLTDD1450 pKa = 3.71YY1451 pKa = 10.99DD1452 pKa = 3.96IIAAAGASFTARR1464 pKa = 11.84TEE1466 pKa = 4.17SFVVNVTDD1474 pKa = 4.24GNLTILFSNITDD1486 pKa = 3.66RR1487 pKa = 11.84AKK1489 pKa = 10.45ISAIEE1494 pKa = 3.91ILGSGNNVAPVIEE1507 pKa = 4.34NPGTQLYY1514 pKa = 10.55SEE1516 pKa = 4.42NLDD1519 pKa = 3.23VNLQMEE1525 pKa = 4.54ATDD1528 pKa = 4.25FNSGDD1533 pKa = 3.56TLTYY1537 pKa = 8.97TAQGLPAALSIDD1549 pKa = 3.6AGTGLISGTLTEE1561 pKa = 4.5PVGTYY1566 pKa = 10.71DD1567 pKa = 3.1VTVRR1571 pKa = 11.84VTDD1574 pKa = 3.19QGGLFDD1580 pKa = 4.95EE1581 pKa = 4.93VTFTIAIGNYY1591 pKa = 8.62TSGLRR1596 pKa = 11.84INAGGAALTHH1606 pKa = 7.25DD1607 pKa = 4.48GNDD1610 pKa = 3.91WIADD1614 pKa = 3.38QYY1616 pKa = 11.64FNGGEE1621 pKa = 4.29LFNSTSPIANTTNDD1635 pKa = 2.63VMYY1638 pKa = 9.29QTEE1641 pKa = 4.51RR1642 pKa = 11.84NSDD1645 pKa = 3.95TPTLSYY1651 pKa = 9.9EE1652 pKa = 4.04IPVPEE1657 pKa = 5.62AGDD1660 pKa = 3.63YY1661 pKa = 10.75AVRR1664 pKa = 11.84LHH1666 pKa = 5.91FAEE1669 pKa = 5.15LFHH1672 pKa = 6.25TAQGQRR1678 pKa = 11.84LFDD1681 pKa = 3.62VDD1683 pKa = 3.79IEE1685 pKa = 4.4SSQVQLTDD1693 pKa = 3.13YY1694 pKa = 10.74DD1695 pKa = 3.79IYY1697 pKa = 11.68VEE1699 pKa = 4.08AGGMNSAVFEE1709 pKa = 4.25TFIVNVTDD1717 pKa = 3.96GALSIVFTNSVDD1729 pKa = 3.3RR1730 pKa = 11.84AKK1732 pKa = 10.65ISGIEE1737 pKa = 3.9VLGEE1741 pKa = 4.06GNFAPILTNPGDD1753 pKa = 3.42QAFDD1757 pKa = 3.29EE1758 pKa = 4.98GASVNLQVEE1767 pKa = 4.27ASDD1770 pKa = 4.09PNAGDD1775 pKa = 3.67VITYY1779 pKa = 9.08SASGLPASLDD1789 pKa = 3.38IDD1791 pKa = 4.11PSTGLISGTLTDD1803 pKa = 4.01AVGDD1807 pKa = 3.72YY1808 pKa = 10.01TVTIRR1813 pKa = 11.84ATDD1816 pKa = 3.5TQGLFDD1822 pKa = 4.42EE1823 pKa = 5.27LSFNITVGTPIRR1835 pKa = 11.84NLLINSGGPSYY1846 pKa = 11.25SFNGEE1851 pKa = 3.85DD1852 pKa = 3.05WSADD1856 pKa = 3.27QSFVGGNTFQVVTPIANTEE1875 pKa = 3.78NDD1877 pKa = 3.02QLYY1880 pKa = 7.53QTEE1883 pKa = 4.5RR1884 pKa = 11.84FGPAPTGSFRR1894 pKa = 11.84YY1895 pKa = 9.65DD1896 pKa = 2.78IPVEE1900 pKa = 4.01NGNYY1904 pKa = 10.37KK1905 pKa = 10.35LDD1907 pKa = 3.51LHH1909 pKa = 6.2FAEE1912 pKa = 5.05IFFGVGTNPGGAGSRR1927 pKa = 11.84IFDD1930 pKa = 3.31VDD1932 pKa = 4.0IEE1934 pKa = 4.29NGQQNLTNYY1943 pKa = 9.89DD1944 pKa = 3.17IVVAAGGSATAIVEE1958 pKa = 4.34SFSNITVSDD1967 pKa = 3.99GSLTIIFTSQVDD1979 pKa = 3.46RR1980 pKa = 11.84AKK1982 pKa = 10.6ISGIQVTSSVPPTVNAGLDD2001 pKa = 3.52QQITLPTNSVTLNGTASDD2019 pKa = 4.29PDD2021 pKa = 3.44GGAILSYY2028 pKa = 10.24QWNQISGPSTATLSGEE2044 pKa = 4.2TTADD2048 pKa = 4.69LIASDD2053 pKa = 4.47LVEE2056 pKa = 4.15GAYY2059 pKa = 10.38VFSLTATDD2067 pKa = 5.04DD2068 pKa = 4.21EE2069 pKa = 5.27NDD2071 pKa = 3.75TASDD2075 pKa = 3.77EE2076 pKa = 4.52VTVTVNPLSGPTALAEE2092 pKa = 4.01ATPLNGTVPLEE2103 pKa = 4.11VTFTGSNSSDD2113 pKa = 3.4DD2114 pKa = 3.77QGVVSYY2120 pKa = 10.78AWDD2123 pKa = 3.84FMDD2126 pKa = 5.1GNTSAEE2132 pKa = 4.12ADD2134 pKa = 3.51PVHH2137 pKa = 7.64IFEE2140 pKa = 4.44TAGTFEE2146 pKa = 4.25VSLTVTDD2153 pKa = 4.5GDD2155 pKa = 4.12NLTDD2159 pKa = 3.89SDD2161 pKa = 4.88TITITVSDD2169 pKa = 4.66PANEE2173 pKa = 4.08APVAVAQADD2182 pKa = 4.19PLSGDD2187 pKa = 3.13IPLEE2191 pKa = 4.08VTFTGSNSTDD2201 pKa = 3.31DD2202 pKa = 4.3KK2203 pKa = 11.4EE2204 pKa = 4.66VVSYY2208 pKa = 10.94AWDD2211 pKa = 3.9FMDD2214 pKa = 5.23GNSAAIADD2222 pKa = 4.14PVHH2225 pKa = 6.28TFTVAGTYY2233 pKa = 9.19EE2234 pKa = 4.13VLLTVTDD2241 pKa = 4.19AEE2243 pKa = 4.85GLTSTATVSITVTEE2257 pKa = 4.4PMNEE2261 pKa = 3.96APVAIALANPTSGQAPLEE2279 pKa = 4.24VTFTGSDD2286 pKa = 3.4STDD2289 pKa = 3.33DD2290 pKa = 3.82NGITTYY2296 pKa = 10.64SWDD2299 pKa = 3.66FMDD2302 pKa = 5.76GSTSSEE2308 pKa = 3.62ADD2310 pKa = 3.45VVHH2313 pKa = 6.63TFTAEE2318 pKa = 3.36GTYY2321 pKa = 10.87NVTLTVTDD2329 pKa = 4.09AEE2331 pKa = 4.55GLTDD2335 pKa = 3.59VAEE2338 pKa = 4.23VTIVVGSAEE2347 pKa = 4.48NLPPTAVIEE2356 pKa = 4.19ATPTTGTVPLEE2367 pKa = 3.76VTFIGRR2373 pKa = 11.84NSQDD2377 pKa = 3.04DD2378 pKa = 3.98FGIVSYY2384 pKa = 11.06NWDD2387 pKa = 3.68FGDD2390 pKa = 4.14GTSSAQIDD2398 pKa = 3.79PMHH2401 pKa = 6.76TFTEE2405 pKa = 4.38SGNYY2409 pKa = 7.75TVSLTVVDD2417 pKa = 4.71AGGLSHH2423 pKa = 7.38TSTITIAVTATSGGMITILLEE2444 pKa = 4.13NPAKK2448 pKa = 10.75DD2449 pKa = 3.58GIARR2453 pKa = 11.84LRR2455 pKa = 11.84VLNKK2459 pKa = 9.98PEE2461 pKa = 3.99DD2462 pKa = 3.81VFVEE2466 pKa = 4.83TIYY2469 pKa = 11.21LHH2471 pKa = 7.06DD2472 pKa = 3.81SSGRR2476 pKa = 11.84LIGGFNAQEE2485 pKa = 3.93IFSDD2489 pKa = 3.26EE2490 pKa = 4.0DD2491 pKa = 3.73AYY2493 pKa = 10.4EE2494 pKa = 4.34VPVNTLRR2501 pKa = 11.84DD2502 pKa = 3.4GLYY2505 pKa = 10.72FIGLEE2510 pKa = 4.08MNEE2513 pKa = 4.01GKK2515 pKa = 10.85AMLVKK2520 pKa = 10.75LLVQNN2525 pKa = 4.6
MM1 pKa = 7.45KK2 pKa = 10.76NKK4 pKa = 8.5ITRR7 pKa = 11.84SNLTPFILNTIPANFTKK24 pKa = 10.34ILAFVIIVFLSNNLLQAQLPQQFQKK49 pKa = 11.05VEE51 pKa = 4.9LITEE55 pKa = 4.01MTNAVNFEE63 pKa = 4.08FAPDD67 pKa = 3.38GRR69 pKa = 11.84IFIVDD74 pKa = 4.0RR75 pKa = 11.84YY76 pKa = 10.97GEE78 pKa = 4.0LLIYY82 pKa = 10.42NPNTQVTVSAGTLEE96 pKa = 4.15VFKK99 pKa = 11.03DD100 pKa = 4.1FEE102 pKa = 6.37DD103 pKa = 4.36GLLGVAFDD111 pKa = 5.62PGFLTNNYY119 pKa = 9.32IYY121 pKa = 10.95LHH123 pKa = 5.72YY124 pKa = 10.87SPLNVSVNRR133 pKa = 11.84VSRR136 pKa = 11.84FTMNGNTLDD145 pKa = 3.91LASEE149 pKa = 4.41VTLMQWPTQRR159 pKa = 11.84NGCCHH164 pKa = 6.72SGGDD168 pKa = 3.99MGFDD172 pKa = 3.57SQGNLYY178 pKa = 10.02IATGDD183 pKa = 3.71NANHH187 pKa = 6.63SDD189 pKa = 3.54YY190 pKa = 11.64AAISEE195 pKa = 4.57TNSVNSAEE203 pKa = 4.35KK204 pKa = 10.66SSSNTNDD211 pKa = 2.95LRR213 pKa = 11.84GKK215 pKa = 9.56ILRR218 pKa = 11.84VRR220 pKa = 11.84PQPDD224 pKa = 3.09GSYY227 pKa = 9.74TIPGGNLFPNGAGGLPEE244 pKa = 4.64IYY246 pKa = 11.06VMGARR251 pKa = 11.84NPYY254 pKa = 10.65RR255 pKa = 11.84MFIDD259 pKa = 4.44KK260 pKa = 10.76EE261 pKa = 4.05NTDD264 pKa = 3.66WIFWGEE270 pKa = 4.14VGPDD274 pKa = 3.21ANVASSAGPEE284 pKa = 4.19GKK286 pKa = 10.12DD287 pKa = 3.53EE288 pKa = 4.52INLTKK293 pKa = 10.22TAGNYY298 pKa = 6.68GWPYY302 pKa = 10.18FSGNNEE308 pKa = 4.35PYY310 pKa = 10.64LIDD313 pKa = 3.6YY314 pKa = 7.72TNPQYY319 pKa = 11.6YY320 pKa = 10.11NDD322 pKa = 4.69PNTPQNVSTWNTGATNLPPARR343 pKa = 11.84PSWLDD348 pKa = 3.53FFHH351 pKa = 7.06KK352 pKa = 10.4SYY354 pKa = 10.27MAGPRR359 pKa = 11.84YY360 pKa = 9.92YY361 pKa = 10.77YY362 pKa = 10.93NPALTDD368 pKa = 3.49QQRR371 pKa = 11.84LPIEE375 pKa = 3.97YY376 pKa = 10.25DD377 pKa = 3.28EE378 pKa = 4.53VFFYY382 pKa = 11.15FDD384 pKa = 4.12FNSSAVWGVKK394 pKa = 8.46MDD396 pKa = 3.57EE397 pKa = 4.05QGIIQSNQRR406 pKa = 11.84LNTAFPANNNLGFIDD421 pKa = 4.13MKK423 pKa = 10.49IGPDD427 pKa = 2.92GHH429 pKa = 8.55LYY431 pKa = 9.7ILEE434 pKa = 4.35YY435 pKa = 10.91GAGCCPNNTGTGKK448 pKa = 10.27LVRR451 pKa = 11.84MDD453 pKa = 3.76YY454 pKa = 10.75IGIVSNSSPVVEE466 pKa = 5.51ISADD470 pKa = 3.63PDD472 pKa = 3.65NGSLPLTVNFSSEE485 pKa = 4.1GTFDD489 pKa = 4.23PDD491 pKa = 3.62GDD493 pKa = 4.17SLTYY497 pKa = 10.24EE498 pKa = 4.17WDD500 pKa = 3.49FEE502 pKa = 4.54TDD504 pKa = 3.21GTVDD508 pKa = 3.37STEE511 pKa = 3.95EE512 pKa = 3.96NPTFVYY518 pKa = 10.75SLAGTYY524 pKa = 9.47NVQLRR529 pKa = 11.84VDD531 pKa = 4.19DD532 pKa = 4.63GNGGVSTKK540 pKa = 10.87NLTIYY545 pKa = 10.44AGNNAPTFTFTSPIDD560 pKa = 3.61GGFMNWGDD568 pKa = 5.75DD569 pKa = 3.24IDD571 pKa = 6.35LDD573 pKa = 4.34LLVEE577 pKa = 4.23DD578 pKa = 4.94VEE580 pKa = 5.69DD581 pKa = 4.47GSTANGGINCLDD593 pKa = 3.81VNVVPSLGHH602 pKa = 6.87LNHH605 pKa = 6.21FHH607 pKa = 7.96DD608 pKa = 6.17DD609 pKa = 3.72MGLDD613 pKa = 4.1GCPKK617 pKa = 9.75TLNLDD622 pKa = 3.9PGNHH626 pKa = 6.53NIYY629 pKa = 11.09GEE631 pKa = 3.81MDD633 pKa = 3.01IFYY636 pKa = 10.85VLGANYY642 pKa = 9.0TDD644 pKa = 3.48QGGLQAFDD652 pKa = 3.33QVQLHH657 pKa = 5.88PKK659 pKa = 9.18RR660 pKa = 11.84RR661 pKa = 11.84EE662 pKa = 3.63AEE664 pKa = 4.19FYY666 pKa = 10.51DD667 pKa = 3.91VKK669 pKa = 11.39SDD671 pKa = 3.2ITVVGNSDD679 pKa = 2.91PWGGGSEE686 pKa = 4.72AIRR689 pKa = 11.84VNHH692 pKa = 6.95DD693 pKa = 3.19GYY695 pKa = 10.79IVFEE699 pKa = 4.13GRR701 pKa = 11.84NLLNISGVKK710 pKa = 9.83YY711 pKa = 10.01RR712 pKa = 11.84VASQTAGGSIEE723 pKa = 4.54LRR725 pKa = 11.84VGSPTGALLATSPVPSTGSLNNWVDD750 pKa = 3.56VEE752 pKa = 4.51SAISDD757 pKa = 3.58PGGKK761 pKa = 9.4NDD763 pKa = 4.6LYY765 pKa = 11.18FVFKK769 pKa = 10.52NASAGVDD776 pKa = 3.21IFDD779 pKa = 4.33LNYY782 pKa = 10.09IEE784 pKa = 6.26FIGQGVSTDD793 pKa = 3.31NTPPVVNNVSSSEE806 pKa = 3.93PTEE809 pKa = 3.83VRR811 pKa = 11.84VEE813 pKa = 3.87FSEE816 pKa = 4.36YY817 pKa = 11.06VNMTVAEE824 pKa = 4.6TITNYY829 pKa = 10.87SLDD832 pKa = 3.54NGEE835 pKa = 4.55SVISASLQEE844 pKa = 4.29DD845 pKa = 3.48QRR847 pKa = 11.84TVILTTSQLAPGITYY862 pKa = 9.12TLDD865 pKa = 3.1INNVTNEE872 pKa = 3.74AGLPVASGSYY882 pKa = 8.75TFNFEE887 pKa = 4.2VEE889 pKa = 4.61CNNTPFPDD897 pKa = 3.09QWQEE901 pKa = 3.84EE902 pKa = 4.83IIGSEE907 pKa = 3.87LPYY910 pKa = 10.31RR911 pKa = 11.84SVYY914 pKa = 10.16IYY916 pKa = 10.88ADD918 pKa = 3.55HH919 pKa = 7.58DD920 pKa = 4.38LDD922 pKa = 6.01GDD924 pKa = 4.03GLKK927 pKa = 10.82DD928 pKa = 3.25IVTGGWWYY936 pKa = 10.82KK937 pKa = 11.09NPGTVTGNWVRR948 pKa = 11.84STIGSPFNNVAYY960 pKa = 10.64VYY962 pKa = 10.87DD963 pKa = 4.06FDD965 pKa = 6.69GDD967 pKa = 3.67GDD969 pKa = 4.27FDD971 pKa = 5.3LLGTQGAYY979 pKa = 9.92EE980 pKa = 4.42GSDD983 pKa = 3.36MAWAQNDD990 pKa = 3.6GSGNFTVYY998 pKa = 10.8TNIDD1002 pKa = 3.7SGQTTYY1008 pKa = 11.04VEE1010 pKa = 4.3PFLAGIAGGVFDD1022 pKa = 4.0NTGDD1026 pKa = 3.59FKK1028 pKa = 11.5LAINWNGAEE1037 pKa = 4.25SSGSPVQLLTIPADD1051 pKa = 3.5PVNTQWTFEE1060 pKa = 4.34NISPDD1065 pKa = 3.56SLGEE1069 pKa = 3.93ALSAGDD1075 pKa = 4.37IDD1077 pKa = 6.36DD1078 pKa = 6.36DD1079 pKa = 5.01GDD1081 pKa = 4.15LDD1083 pKa = 5.28LFQGSNWLRR1092 pKa = 11.84NEE1094 pKa = 4.37GNGTWTTFSTGITYY1108 pKa = 7.76PTTMDD1113 pKa = 3.45RR1114 pKa = 11.84NMLADD1119 pKa = 4.43FDD1121 pKa = 5.32RR1122 pKa = 11.84DD1123 pKa = 3.33GDD1125 pKa = 3.8LDD1127 pKa = 3.94AVVSQLGSNQEE1138 pKa = 4.1IAWFEE1143 pKa = 3.97APEE1146 pKa = 5.18DD1147 pKa = 3.91PTQPWTKK1154 pKa = 9.24NTLDD1158 pKa = 3.45TDD1160 pKa = 3.42IDD1162 pKa = 3.91GGLSVDD1168 pKa = 4.69VIDD1171 pKa = 5.94FDD1173 pKa = 5.11FDD1175 pKa = 3.6GDD1177 pKa = 3.56ADD1179 pKa = 3.73IVVGEE1184 pKa = 4.16WQSAYY1189 pKa = 10.94RR1190 pKa = 11.84LFAFEE1195 pKa = 5.71NDD1197 pKa = 3.48LCNTGTWIKK1206 pKa = 9.26HH1207 pKa = 5.38TIHH1210 pKa = 7.24PGTPTFDD1217 pKa = 3.38HH1218 pKa = 7.21HH1219 pKa = 7.72DD1220 pKa = 3.69GAQVVDD1226 pKa = 3.55IDD1228 pKa = 4.28EE1229 pKa = 5.94DD1230 pKa = 4.33GDD1232 pKa = 4.22LDD1234 pKa = 3.87IVSIGWNNITPRR1246 pKa = 11.84IFEE1249 pKa = 4.31NTTGSAPVNSPPMVLNPGDD1268 pKa = 3.43QLFNEE1273 pKa = 4.71GAIVNLQIQGSDD1285 pKa = 3.35PDD1287 pKa = 4.02FSDD1290 pKa = 3.17VLTYY1294 pKa = 9.03TAQGLPADD1302 pKa = 4.77LSIDD1306 pKa = 3.89PDD1308 pKa = 3.64TGIIGGNLSAAQGSYY1323 pKa = 10.15PVTVRR1328 pKa = 11.84VTDD1331 pKa = 3.32NGGLFDD1337 pKa = 4.17EE1338 pKa = 4.88TTFTIFVGAFNSIIRR1353 pKa = 11.84INAGGPLVTVNNEE1366 pKa = 3.24DD1367 pKa = 3.89WIADD1371 pKa = 3.41QYY1373 pKa = 11.56FNGGATFSTANAINGTTDD1391 pKa = 4.22DD1392 pKa = 4.04ILYY1395 pKa = 8.01QTEE1398 pKa = 4.36RR1399 pKa = 11.84NSDD1402 pKa = 4.05DD1403 pKa = 3.26PTLTYY1408 pKa = 10.02QIPVPEE1414 pKa = 4.19NGEE1417 pKa = 3.98YY1418 pKa = 10.63GVRR1421 pKa = 11.84LLFAEE1426 pKa = 4.68IFHH1429 pKa = 6.7NSTGSRR1435 pKa = 11.84LFDD1438 pKa = 3.36VDD1440 pKa = 3.35IEE1442 pKa = 4.52GGQGQLTDD1450 pKa = 3.71YY1451 pKa = 10.99DD1452 pKa = 3.96IIAAAGASFTARR1464 pKa = 11.84TEE1466 pKa = 4.17SFVVNVTDD1474 pKa = 4.24GNLTILFSNITDD1486 pKa = 3.66RR1487 pKa = 11.84AKK1489 pKa = 10.45ISAIEE1494 pKa = 3.91ILGSGNNVAPVIEE1507 pKa = 4.34NPGTQLYY1514 pKa = 10.55SEE1516 pKa = 4.42NLDD1519 pKa = 3.23VNLQMEE1525 pKa = 4.54ATDD1528 pKa = 4.25FNSGDD1533 pKa = 3.56TLTYY1537 pKa = 8.97TAQGLPAALSIDD1549 pKa = 3.6AGTGLISGTLTEE1561 pKa = 4.5PVGTYY1566 pKa = 10.71DD1567 pKa = 3.1VTVRR1571 pKa = 11.84VTDD1574 pKa = 3.19QGGLFDD1580 pKa = 4.95EE1581 pKa = 4.93VTFTIAIGNYY1591 pKa = 8.62TSGLRR1596 pKa = 11.84INAGGAALTHH1606 pKa = 7.25DD1607 pKa = 4.48GNDD1610 pKa = 3.91WIADD1614 pKa = 3.38QYY1616 pKa = 11.64FNGGEE1621 pKa = 4.29LFNSTSPIANTTNDD1635 pKa = 2.63VMYY1638 pKa = 9.29QTEE1641 pKa = 4.51RR1642 pKa = 11.84NSDD1645 pKa = 3.95TPTLSYY1651 pKa = 9.9EE1652 pKa = 4.04IPVPEE1657 pKa = 5.62AGDD1660 pKa = 3.63YY1661 pKa = 10.75AVRR1664 pKa = 11.84LHH1666 pKa = 5.91FAEE1669 pKa = 5.15LFHH1672 pKa = 6.25TAQGQRR1678 pKa = 11.84LFDD1681 pKa = 3.62VDD1683 pKa = 3.79IEE1685 pKa = 4.4SSQVQLTDD1693 pKa = 3.13YY1694 pKa = 10.74DD1695 pKa = 3.79IYY1697 pKa = 11.68VEE1699 pKa = 4.08AGGMNSAVFEE1709 pKa = 4.25TFIVNVTDD1717 pKa = 3.96GALSIVFTNSVDD1729 pKa = 3.3RR1730 pKa = 11.84AKK1732 pKa = 10.65ISGIEE1737 pKa = 3.9VLGEE1741 pKa = 4.06GNFAPILTNPGDD1753 pKa = 3.42QAFDD1757 pKa = 3.29EE1758 pKa = 4.98GASVNLQVEE1767 pKa = 4.27ASDD1770 pKa = 4.09PNAGDD1775 pKa = 3.67VITYY1779 pKa = 9.08SASGLPASLDD1789 pKa = 3.38IDD1791 pKa = 4.11PSTGLISGTLTDD1803 pKa = 4.01AVGDD1807 pKa = 3.72YY1808 pKa = 10.01TVTIRR1813 pKa = 11.84ATDD1816 pKa = 3.5TQGLFDD1822 pKa = 4.42EE1823 pKa = 5.27LSFNITVGTPIRR1835 pKa = 11.84NLLINSGGPSYY1846 pKa = 11.25SFNGEE1851 pKa = 3.85DD1852 pKa = 3.05WSADD1856 pKa = 3.27QSFVGGNTFQVVTPIANTEE1875 pKa = 3.78NDD1877 pKa = 3.02QLYY1880 pKa = 7.53QTEE1883 pKa = 4.5RR1884 pKa = 11.84FGPAPTGSFRR1894 pKa = 11.84YY1895 pKa = 9.65DD1896 pKa = 2.78IPVEE1900 pKa = 4.01NGNYY1904 pKa = 10.37KK1905 pKa = 10.35LDD1907 pKa = 3.51LHH1909 pKa = 6.2FAEE1912 pKa = 5.05IFFGVGTNPGGAGSRR1927 pKa = 11.84IFDD1930 pKa = 3.31VDD1932 pKa = 4.0IEE1934 pKa = 4.29NGQQNLTNYY1943 pKa = 9.89DD1944 pKa = 3.17IVVAAGGSATAIVEE1958 pKa = 4.34SFSNITVSDD1967 pKa = 3.99GSLTIIFTSQVDD1979 pKa = 3.46RR1980 pKa = 11.84AKK1982 pKa = 10.6ISGIQVTSSVPPTVNAGLDD2001 pKa = 3.52QQITLPTNSVTLNGTASDD2019 pKa = 4.29PDD2021 pKa = 3.44GGAILSYY2028 pKa = 10.24QWNQISGPSTATLSGEE2044 pKa = 4.2TTADD2048 pKa = 4.69LIASDD2053 pKa = 4.47LVEE2056 pKa = 4.15GAYY2059 pKa = 10.38VFSLTATDD2067 pKa = 5.04DD2068 pKa = 4.21EE2069 pKa = 5.27NDD2071 pKa = 3.75TASDD2075 pKa = 3.77EE2076 pKa = 4.52VTVTVNPLSGPTALAEE2092 pKa = 4.01ATPLNGTVPLEE2103 pKa = 4.11VTFTGSNSSDD2113 pKa = 3.4DD2114 pKa = 3.77QGVVSYY2120 pKa = 10.78AWDD2123 pKa = 3.84FMDD2126 pKa = 5.1GNTSAEE2132 pKa = 4.12ADD2134 pKa = 3.51PVHH2137 pKa = 7.64IFEE2140 pKa = 4.44TAGTFEE2146 pKa = 4.25VSLTVTDD2153 pKa = 4.5GDD2155 pKa = 4.12NLTDD2159 pKa = 3.89SDD2161 pKa = 4.88TITITVSDD2169 pKa = 4.66PANEE2173 pKa = 4.08APVAVAQADD2182 pKa = 4.19PLSGDD2187 pKa = 3.13IPLEE2191 pKa = 4.08VTFTGSNSTDD2201 pKa = 3.31DD2202 pKa = 4.3KK2203 pKa = 11.4EE2204 pKa = 4.66VVSYY2208 pKa = 10.94AWDD2211 pKa = 3.9FMDD2214 pKa = 5.23GNSAAIADD2222 pKa = 4.14PVHH2225 pKa = 6.28TFTVAGTYY2233 pKa = 9.19EE2234 pKa = 4.13VLLTVTDD2241 pKa = 4.19AEE2243 pKa = 4.85GLTSTATVSITVTEE2257 pKa = 4.4PMNEE2261 pKa = 3.96APVAIALANPTSGQAPLEE2279 pKa = 4.24VTFTGSDD2286 pKa = 3.4STDD2289 pKa = 3.33DD2290 pKa = 3.82NGITTYY2296 pKa = 10.64SWDD2299 pKa = 3.66FMDD2302 pKa = 5.76GSTSSEE2308 pKa = 3.62ADD2310 pKa = 3.45VVHH2313 pKa = 6.63TFTAEE2318 pKa = 3.36GTYY2321 pKa = 10.87NVTLTVTDD2329 pKa = 4.09AEE2331 pKa = 4.55GLTDD2335 pKa = 3.59VAEE2338 pKa = 4.23VTIVVGSAEE2347 pKa = 4.48NLPPTAVIEE2356 pKa = 4.19ATPTTGTVPLEE2367 pKa = 3.76VTFIGRR2373 pKa = 11.84NSQDD2377 pKa = 3.04DD2378 pKa = 3.98FGIVSYY2384 pKa = 11.06NWDD2387 pKa = 3.68FGDD2390 pKa = 4.14GTSSAQIDD2398 pKa = 3.79PMHH2401 pKa = 6.76TFTEE2405 pKa = 4.38SGNYY2409 pKa = 7.75TVSLTVVDD2417 pKa = 4.71AGGLSHH2423 pKa = 7.38TSTITIAVTATSGGMITILLEE2444 pKa = 4.13NPAKK2448 pKa = 10.75DD2449 pKa = 3.58GIARR2453 pKa = 11.84LRR2455 pKa = 11.84VLNKK2459 pKa = 9.98PEE2461 pKa = 3.99DD2462 pKa = 3.81VFVEE2466 pKa = 4.83TIYY2469 pKa = 11.21LHH2471 pKa = 7.06DD2472 pKa = 3.81SSGRR2476 pKa = 11.84LIGGFNAQEE2485 pKa = 3.93IFSDD2489 pKa = 3.26EE2490 pKa = 4.0DD2491 pKa = 3.73AYY2493 pKa = 10.4EE2494 pKa = 4.34VPVNTLRR2501 pKa = 11.84DD2502 pKa = 3.4GLYY2505 pKa = 10.72FIGLEE2510 pKa = 4.08MNEE2513 pKa = 4.01GKK2515 pKa = 10.85AMLVKK2520 pKa = 10.75LLVQNN2525 pKa = 4.6
Molecular weight: 271.01 kDa
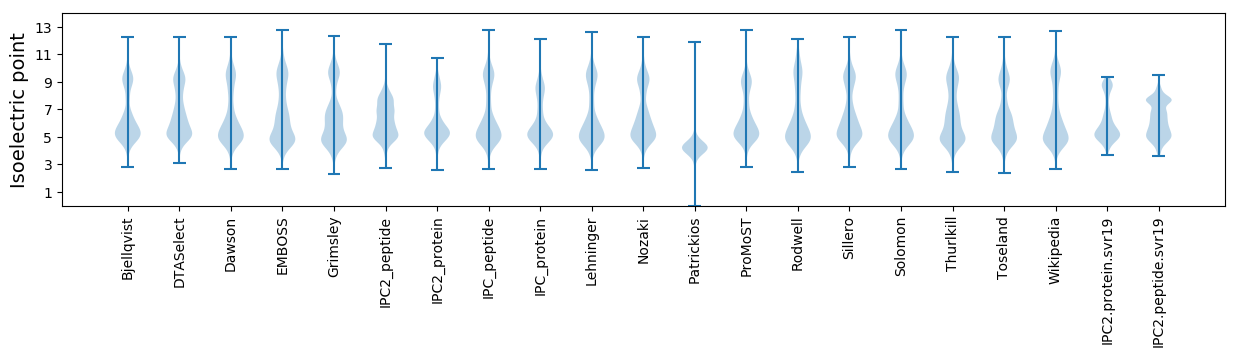
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6TMK9|A0A4R6TMK9_9FLAO Uncharacterized protein OS=Zeaxanthinibacter enoshimensis OX=392009 GN=CLV82_0273 PE=4 SV=1
MM1 pKa = 8.13RR2 pKa = 11.84ILIIGGTGKK11 pKa = 8.69TGRR14 pKa = 11.84EE15 pKa = 4.11LIKK18 pKa = 10.51QGLAKK23 pKa = 10.36GHH25 pKa = 6.73TITVFARR32 pKa = 11.84KK33 pKa = 8.52PGKK36 pKa = 9.76IRR38 pKa = 11.84EE39 pKa = 4.01VHH41 pKa = 5.22QNLRR45 pKa = 11.84VVPGNILDD53 pKa = 4.0PTTLAPALLNQDD65 pKa = 3.6AVLSALGHH73 pKa = 5.47KK74 pKa = 10.18RR75 pKa = 11.84FVIRR79 pKa = 11.84TNTLSEE85 pKa = 4.14GTKK88 pKa = 10.5NLISGMEE95 pKa = 3.8QLGVKK100 pKa = 10.13RR101 pKa = 11.84LICITSLGIADD112 pKa = 3.33SRR114 pKa = 11.84YY115 pKa = 10.12RR116 pKa = 11.84LGLYY120 pKa = 10.12YY121 pKa = 10.57SLFVIPVLLFFYY133 pKa = 10.63FRR135 pKa = 11.84DD136 pKa = 3.49KK137 pKa = 10.95EE138 pKa = 4.04KK139 pKa = 10.16QEE141 pKa = 3.88RR142 pKa = 11.84VIRR145 pKa = 11.84EE146 pKa = 4.16SNLDD150 pKa = 3.29WTIVRR155 pKa = 11.84PGQLTNGKK163 pKa = 9.45KK164 pKa = 7.62RR165 pKa = 11.84TRR167 pKa = 11.84YY168 pKa = 7.91RR169 pKa = 11.84TGPGLGHH176 pKa = 6.67YY177 pKa = 10.04IFTRR181 pKa = 11.84LVSRR185 pKa = 11.84ATVAGFMLDD194 pKa = 3.33EE195 pKa = 4.35LAGRR199 pKa = 11.84NYY201 pKa = 10.29LKK203 pKa = 10.33QVSGITNN210 pKa = 3.82
MM1 pKa = 8.13RR2 pKa = 11.84ILIIGGTGKK11 pKa = 8.69TGRR14 pKa = 11.84EE15 pKa = 4.11LIKK18 pKa = 10.51QGLAKK23 pKa = 10.36GHH25 pKa = 6.73TITVFARR32 pKa = 11.84KK33 pKa = 8.52PGKK36 pKa = 9.76IRR38 pKa = 11.84EE39 pKa = 4.01VHH41 pKa = 5.22QNLRR45 pKa = 11.84VVPGNILDD53 pKa = 4.0PTTLAPALLNQDD65 pKa = 3.6AVLSALGHH73 pKa = 5.47KK74 pKa = 10.18RR75 pKa = 11.84FVIRR79 pKa = 11.84TNTLSEE85 pKa = 4.14GTKK88 pKa = 10.5NLISGMEE95 pKa = 3.8QLGVKK100 pKa = 10.13RR101 pKa = 11.84LICITSLGIADD112 pKa = 3.33SRR114 pKa = 11.84YY115 pKa = 10.12RR116 pKa = 11.84LGLYY120 pKa = 10.12YY121 pKa = 10.57SLFVIPVLLFFYY133 pKa = 10.63FRR135 pKa = 11.84DD136 pKa = 3.49KK137 pKa = 10.95EE138 pKa = 4.04KK139 pKa = 10.16QEE141 pKa = 3.88RR142 pKa = 11.84VIRR145 pKa = 11.84EE146 pKa = 4.16SNLDD150 pKa = 3.29WTIVRR155 pKa = 11.84PGQLTNGKK163 pKa = 9.45KK164 pKa = 7.62RR165 pKa = 11.84TRR167 pKa = 11.84YY168 pKa = 7.91RR169 pKa = 11.84TGPGLGHH176 pKa = 6.67YY177 pKa = 10.04IFTRR181 pKa = 11.84LVSRR185 pKa = 11.84ATVAGFMLDD194 pKa = 3.33EE195 pKa = 4.35LAGRR199 pKa = 11.84NYY201 pKa = 10.29LKK203 pKa = 10.33QVSGITNN210 pKa = 3.82
Molecular weight: 23.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007827 |
28 |
4479 |
344.4 |
38.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.074 ± 0.04 | 0.741 ± 0.018 |
5.876 ± 0.062 | 7.062 ± 0.048 |
4.819 ± 0.035 | 7.181 ± 0.06 |
1.906 ± 0.03 | 6.853 ± 0.039 |
6.32 ± 0.072 | 9.697 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.027 | 4.917 ± 0.049 |
3.919 ± 0.03 | 3.585 ± 0.029 |
4.474 ± 0.039 | 6.167 ± 0.037 |
5.492 ± 0.073 | 6.414 ± 0.029 |
1.111 ± 0.02 | 3.965 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
