
Microbacterium phage Anakin
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Krampusvirus; unclassified Krampusvirus
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
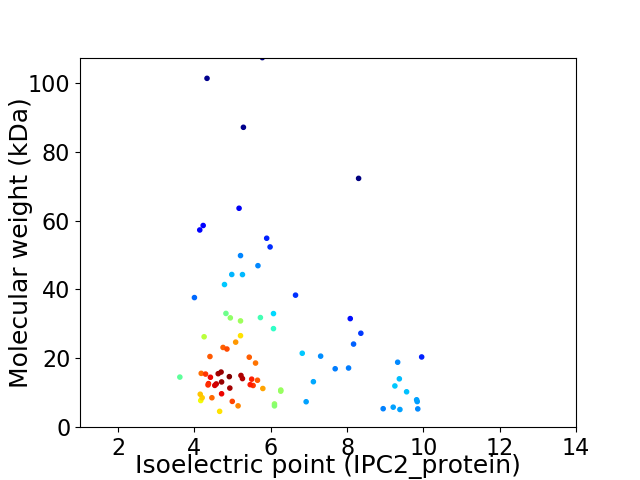
Virtual 2D-PAGE plot for 82 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y6EH94|A0A4Y6EH94_9CAUD Uncharacterized protein OS=Microbacterium phage Anakin OX=2588136 GN=41 PE=4 SV=1
MM1 pKa = 7.68NIRR4 pKa = 11.84RR5 pKa = 11.84LVATAATALVVGGGLAVGTALPASAHH31 pKa = 5.54TPSVSATCSAIDD43 pKa = 3.57INLTNYY49 pKa = 7.61EE50 pKa = 4.31VKK52 pKa = 10.33PEE54 pKa = 4.18SGTPTIEE61 pKa = 4.0IANPDD66 pKa = 4.07YY67 pKa = 10.99VPEE70 pKa = 4.28VPGVPAQGTPTILVPNPDD88 pKa = 4.11YY89 pKa = 11.35VPAIPEE95 pKa = 4.04VPEE98 pKa = 4.03VLGDD102 pKa = 3.62PPLIKK107 pKa = 10.41AEE109 pKa = 3.92QLEE112 pKa = 4.61VSHH115 pKa = 6.64TEE117 pKa = 3.65NEE119 pKa = 4.08YY120 pKa = 10.46KK121 pKa = 10.31QWVTGKK127 pKa = 10.14LKK129 pKa = 9.7WVKK132 pKa = 10.71SDD134 pKa = 3.14DD135 pKa = 3.65WNPGFGWYY143 pKa = 8.02ATGNQRR149 pKa = 11.84IVVDD153 pKa = 4.0VPYY156 pKa = 10.73SPAVYY161 pKa = 10.01GPQPVITSYY170 pKa = 11.04QPAVPAVGEE179 pKa = 4.02PQIEE183 pKa = 4.56VANPAYY189 pKa = 10.37VPAVPAVPAVGEE201 pKa = 4.0PTKK204 pKa = 10.47TVEE207 pKa = 4.04NPDD210 pKa = 3.5YY211 pKa = 10.7VAADD215 pKa = 3.55ATPNTVTVIVDD226 pKa = 3.45GEE228 pKa = 4.34EE229 pKa = 4.06VLDD232 pKa = 4.01EE233 pKa = 4.4EE234 pKa = 5.69FGTSYY239 pKa = 11.18SNSVAIDD246 pKa = 3.3GTKK249 pKa = 8.57NHH251 pKa = 6.43SYY253 pKa = 9.87KK254 pKa = 10.52VEE256 pKa = 4.05VVGYY260 pKa = 8.19TGVGTKK266 pKa = 9.28TFTGKK271 pKa = 7.2TTACPPTPVVAPTLTVLPPTCDD293 pKa = 3.4ADD295 pKa = 3.78GSLPFLGNPAAQNPNGYY312 pKa = 8.88EE313 pKa = 4.09FPGQGYY319 pKa = 8.68RR320 pKa = 11.84VYY322 pKa = 10.82LDD324 pKa = 3.47KK325 pKa = 11.43SFTGAGTYY333 pKa = 9.78VATIQKK339 pKa = 9.52VGPGFDD345 pKa = 3.18PAFPYY350 pKa = 8.35GTKK353 pKa = 10.4VSGEE357 pKa = 4.25TKK359 pKa = 8.92QTLTVLPATGYY370 pKa = 10.61QGTDD374 pKa = 3.31PEE376 pKa = 4.64APCYY380 pKa = 10.8VPVPEE385 pKa = 4.23NTRR388 pKa = 11.84EE389 pKa = 4.1DD390 pKa = 4.66GEE392 pKa = 4.29WSTPIITCEE401 pKa = 4.0NEE403 pKa = 3.79VGDD406 pKa = 4.8EE407 pKa = 3.87ITITRR412 pKa = 11.84EE413 pKa = 3.82VTFTEE418 pKa = 4.17HH419 pKa = 6.15TLNTEE424 pKa = 3.91TGKK427 pKa = 10.25VEE429 pKa = 4.07TTTEE433 pKa = 4.08VVNEE437 pKa = 3.73NDD439 pKa = 3.44VYY441 pKa = 10.93IVTEE445 pKa = 3.87ADD447 pKa = 3.28IAEE450 pKa = 4.51LDD452 pKa = 4.18CPVVTPEE459 pKa = 3.97EE460 pKa = 4.51PEE462 pKa = 4.29TPSEE466 pKa = 4.13EE467 pKa = 4.43PEE469 pKa = 4.55TPTEE473 pKa = 4.24TPNAPTEE480 pKa = 4.47GNGTTLRR487 pKa = 11.84AATTQNDD494 pKa = 4.28AEE496 pKa = 4.45TLAQTGADD504 pKa = 4.08SPLPWVLGGTSLALGGLALWLFGIYY529 pKa = 9.61RR530 pKa = 11.84RR531 pKa = 11.84NQGGVVRR538 pKa = 11.84SGNPEE543 pKa = 3.47
MM1 pKa = 7.68NIRR4 pKa = 11.84RR5 pKa = 11.84LVATAATALVVGGGLAVGTALPASAHH31 pKa = 5.54TPSVSATCSAIDD43 pKa = 3.57INLTNYY49 pKa = 7.61EE50 pKa = 4.31VKK52 pKa = 10.33PEE54 pKa = 4.18SGTPTIEE61 pKa = 4.0IANPDD66 pKa = 4.07YY67 pKa = 10.99VPEE70 pKa = 4.28VPGVPAQGTPTILVPNPDD88 pKa = 4.11YY89 pKa = 11.35VPAIPEE95 pKa = 4.04VPEE98 pKa = 4.03VLGDD102 pKa = 3.62PPLIKK107 pKa = 10.41AEE109 pKa = 3.92QLEE112 pKa = 4.61VSHH115 pKa = 6.64TEE117 pKa = 3.65NEE119 pKa = 4.08YY120 pKa = 10.46KK121 pKa = 10.31QWVTGKK127 pKa = 10.14LKK129 pKa = 9.7WVKK132 pKa = 10.71SDD134 pKa = 3.14DD135 pKa = 3.65WNPGFGWYY143 pKa = 8.02ATGNQRR149 pKa = 11.84IVVDD153 pKa = 4.0VPYY156 pKa = 10.73SPAVYY161 pKa = 10.01GPQPVITSYY170 pKa = 11.04QPAVPAVGEE179 pKa = 4.02PQIEE183 pKa = 4.56VANPAYY189 pKa = 10.37VPAVPAVPAVGEE201 pKa = 4.0PTKK204 pKa = 10.47TVEE207 pKa = 4.04NPDD210 pKa = 3.5YY211 pKa = 10.7VAADD215 pKa = 3.55ATPNTVTVIVDD226 pKa = 3.45GEE228 pKa = 4.34EE229 pKa = 4.06VLDD232 pKa = 4.01EE233 pKa = 4.4EE234 pKa = 5.69FGTSYY239 pKa = 11.18SNSVAIDD246 pKa = 3.3GTKK249 pKa = 8.57NHH251 pKa = 6.43SYY253 pKa = 9.87KK254 pKa = 10.52VEE256 pKa = 4.05VVGYY260 pKa = 8.19TGVGTKK266 pKa = 9.28TFTGKK271 pKa = 7.2TTACPPTPVVAPTLTVLPPTCDD293 pKa = 3.4ADD295 pKa = 3.78GSLPFLGNPAAQNPNGYY312 pKa = 8.88EE313 pKa = 4.09FPGQGYY319 pKa = 8.68RR320 pKa = 11.84VYY322 pKa = 10.82LDD324 pKa = 3.47KK325 pKa = 11.43SFTGAGTYY333 pKa = 9.78VATIQKK339 pKa = 9.52VGPGFDD345 pKa = 3.18PAFPYY350 pKa = 8.35GTKK353 pKa = 10.4VSGEE357 pKa = 4.25TKK359 pKa = 8.92QTLTVLPATGYY370 pKa = 10.61QGTDD374 pKa = 3.31PEE376 pKa = 4.64APCYY380 pKa = 10.8VPVPEE385 pKa = 4.23NTRR388 pKa = 11.84EE389 pKa = 4.1DD390 pKa = 4.66GEE392 pKa = 4.29WSTPIITCEE401 pKa = 4.0NEE403 pKa = 3.79VGDD406 pKa = 4.8EE407 pKa = 3.87ITITRR412 pKa = 11.84EE413 pKa = 3.82VTFTEE418 pKa = 4.17HH419 pKa = 6.15TLNTEE424 pKa = 3.91TGKK427 pKa = 10.25VEE429 pKa = 4.07TTTEE433 pKa = 4.08VVNEE437 pKa = 3.73NDD439 pKa = 3.44VYY441 pKa = 10.93IVTEE445 pKa = 3.87ADD447 pKa = 3.28IAEE450 pKa = 4.51LDD452 pKa = 4.18CPVVTPEE459 pKa = 3.97EE460 pKa = 4.51PEE462 pKa = 4.29TPSEE466 pKa = 4.13EE467 pKa = 4.43PEE469 pKa = 4.55TPTEE473 pKa = 4.24TPNAPTEE480 pKa = 4.47GNGTTLRR487 pKa = 11.84AATTQNDD494 pKa = 4.28AEE496 pKa = 4.45TLAQTGADD504 pKa = 4.08SPLPWVLGGTSLALGGLALWLFGIYY529 pKa = 9.61RR530 pKa = 11.84RR531 pKa = 11.84NQGGVVRR538 pKa = 11.84SGNPEE543 pKa = 3.47
Molecular weight: 57.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y6ES99|A0A4Y6ES99_9CAUD Uncharacterized protein OS=Microbacterium phage Anakin OX=2588136 GN=2 PE=4 SV=1
MM1 pKa = 7.89PLLKK5 pKa = 9.67TLPTKK10 pKa = 10.42PGAYY14 pKa = 8.99MSSKK18 pKa = 10.82DD19 pKa = 3.33RR20 pKa = 11.84GIWLLEE26 pKa = 4.08AKK28 pKa = 10.26SDD30 pKa = 3.54GGLYY34 pKa = 9.61WRR36 pKa = 11.84MPGGFLMIDD45 pKa = 3.49SAARR49 pKa = 11.84SGPMSKK55 pKa = 10.08VVAEE59 pKa = 4.27HH60 pKa = 6.42LPFRR64 pKa = 11.84RR65 pKa = 11.84MLPSRR70 pKa = 11.84RR71 pKa = 3.69
MM1 pKa = 7.89PLLKK5 pKa = 9.67TLPTKK10 pKa = 10.42PGAYY14 pKa = 8.99MSSKK18 pKa = 10.82DD19 pKa = 3.33RR20 pKa = 11.84GIWLLEE26 pKa = 4.08AKK28 pKa = 10.26SDD30 pKa = 3.54GGLYY34 pKa = 9.61WRR36 pKa = 11.84MPGGFLMIDD45 pKa = 3.49SAARR49 pKa = 11.84SGPMSKK55 pKa = 10.08VVAEE59 pKa = 4.27HH60 pKa = 6.42LPFRR64 pKa = 11.84RR65 pKa = 11.84MLPSRR70 pKa = 11.84RR71 pKa = 3.69
Molecular weight: 7.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17976 |
40 |
961 |
219.2 |
24.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.553 ± 0.482 | 0.729 ± 0.115 |
6.815 ± 0.322 | 7.699 ± 0.386 |
3.149 ± 0.164 | 7.766 ± 0.217 |
1.836 ± 0.138 | 4.918 ± 0.261 |
5.174 ± 0.296 | 7.304 ± 0.233 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.559 ± 0.148 | 3.538 ± 0.233 |
4.918 ± 0.298 | 3.56 ± 0.161 |
6.197 ± 0.373 | 5.357 ± 0.19 |
6.364 ± 0.332 | 6.653 ± 0.294 |
2.008 ± 0.151 | 2.904 ± 0.186 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
